Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
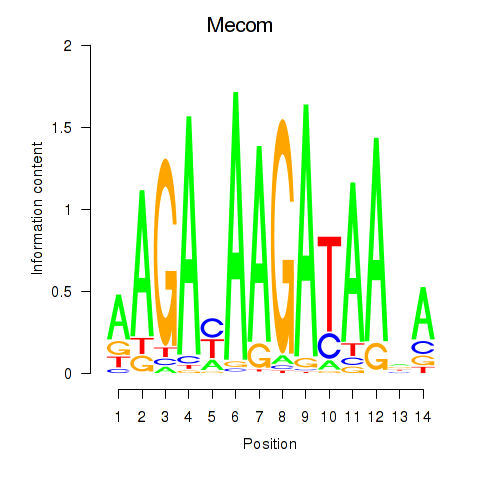
Results for Mecom
Z-value: 0.26
Transcription factors associated with Mecom
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mecom
|
ENSRNOG00000012645 | MDS1 and EVI1 complex locus |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mecom | rn6_v1_chr2_-_117454769_117454769 | -0.48 | 4.2e-01 | Click! |
Activity profile of Mecom motif
Sorted Z-values of Mecom motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_50381244 | 0.11 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr1_-_20155960 | 0.07 |
ENSRNOT00000061389
|
Samd3
|
sterile alpha motif domain containing 3 |
| chr8_+_14060394 | 0.06 |
ENSRNOT00000014827
|
Smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr5_+_173148884 | 0.06 |
ENSRNOT00000041753
|
LOC100364191
|
hCG1994130-like |
| chr16_+_75966352 | 0.05 |
ENSRNOT00000022774
|
Angpt2
|
angiopoietin 2 |
| chr2_-_77784522 | 0.05 |
ENSRNOT00000030328
|
LOC310177
|
similar to RIKEN cDNA 0610040D20 |
| chr17_+_11101306 | 0.05 |
ENSRNOT00000030893
|
Drd1
|
dopamine receptor D1 |
| chr8_+_71514281 | 0.05 |
ENSRNOT00000022256
|
Ns5atp9
|
NS5A (hepatitis C virus) transactivated protein 9 |
| chr16_-_82439441 | 0.05 |
ENSRNOT00000040315
|
AABR07026539.1
|
|
| chr2_-_256154584 | 0.05 |
ENSRNOT00000072487
|
AABR07013776.1
|
|
| chr1_+_93242050 | 0.04 |
ENSRNOT00000013741
|
RGD1562402
|
similar to 60S ribosomal protein L27a |
| chr7_+_12471824 | 0.04 |
ENSRNOT00000068197
|
Sbno2
|
strawberry notch homolog 2 |
| chr6_+_99356509 | 0.03 |
ENSRNOT00000008416
|
Akap5
|
A-kinase anchoring protein 5 |
| chr1_+_42121636 | 0.03 |
ENSRNOT00000025616
|
Myct1
|
myc target 1 |
| chr10_-_66873948 | 0.03 |
ENSRNOT00000039261
|
Evi2a
|
ecotropic viral integration site 2A |
| chr9_-_76768770 | 0.03 |
ENSRNOT00000087779
ENSRNOT00000057849 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr3_-_148312420 | 0.03 |
ENSRNOT00000047416
ENSRNOT00000081272 |
Bcl2l1
|
Bcl2-like 1 |
| chr13_-_107831014 | 0.03 |
ENSRNOT00000003684
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr3_-_107760550 | 0.03 |
ENSRNOT00000077091
ENSRNOT00000051638 |
Meis2
|
Meis homeobox 2 |
| chr11_+_64601029 | 0.02 |
ENSRNOT00000004138
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr3_-_44177689 | 0.02 |
ENSRNOT00000006387
|
Cytip
|
cytohesin 1 interacting protein |
| chr4_-_131694755 | 0.02 |
ENSRNOT00000013271
|
Foxp1
|
forkhead box P1 |
| chr5_+_154265097 | 0.02 |
ENSRNOT00000012342
|
Cnr2
|
cannabinoid receptor 2 |
| chr9_-_26932201 | 0.02 |
ENSRNOT00000017081
|
Mcm3
|
minichromosome maintenance complex component 3 |
| chr12_+_41636994 | 0.01 |
ENSRNOT00000001882
|
Sdsl
|
serine dehydratase-like |
| chr14_+_63095720 | 0.01 |
ENSRNOT00000006071
|
Ppargc1a
|
PPARG coactivator 1 alpha |
| chr10_+_57251253 | 0.01 |
ENSRNOT00000043171
|
LOC108348287
|
60S ribosomal protein L36a |
| chr1_-_167765552 | 0.01 |
ENSRNOT00000024954
|
Bcl2l1-ps1
|
Bcl2-like 1, pseudogene 1 |
| chr11_+_44877859 | 0.01 |
ENSRNOT00000060838
|
Col8a1
|
collagen type VIII alpha 1 chain |
| chr2_-_196582997 | 0.01 |
ENSRNOT00000081672
|
AABR07012475.1
|
|
| chr20_+_1787174 | 0.01 |
ENSRNOT00000072824
ENSRNOT00000041883 |
Olr1738
|
olfactory receptor 1738 |
| chr5_+_69851171 | 0.01 |
ENSRNOT00000047780
|
AABR07048271.1
|
|
| chr1_-_93949187 | 0.01 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr1_+_128924966 | 0.01 |
ENSRNOT00000019267
|
Igf1r
|
insulin-like growth factor 1 receptor |
| chr2_-_3124543 | 0.01 |
ENSRNOT00000036547
|
Fam81b
|
family with sequence similarity 81, member B |
| chr4_-_58250798 | 0.01 |
ENSRNOT00000048436
|
Klf14
|
Kruppel-like factor 14 |
| chr13_+_89826254 | 0.01 |
ENSRNOT00000006141
|
F11r
|
F11 receptor |
| chr2_-_187786700 | 0.01 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr19_+_38846201 | 0.01 |
ENSRNOT00000030768
ENSRNOT00000063977 |
Tango6
|
transport and golgi organization 6 homolog |
| chr15_-_27726646 | 0.01 |
ENSRNOT00000082139
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr8_+_113603533 | 0.01 |
ENSRNOT00000017280
|
Mrpl3
|
mitochondrial ribosomal protein L3 |
| chrX_+_21696772 | 0.01 |
ENSRNOT00000043559
|
Hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr4_+_142780987 | 0.01 |
ENSRNOT00000056570
|
Grm7
|
glutamate metabotropic receptor 7 |
| chr3_-_4341771 | 0.00 |
ENSRNOT00000034694
|
LOC684988
|
similar to ribosomal protein S13 |
| chr11_+_60383431 | 0.00 |
ENSRNOT00000093295
|
Cd200
|
Cd200 molecule |
| chr13_-_35048444 | 0.00 |
ENSRNOT00000009963
|
Gli2
|
GLI family zinc finger 2 |
| chr18_-_5314511 | 0.00 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr19_-_38344845 | 0.00 |
ENSRNOT00000072666
|
AABR07043748.1
|
|
| chr10_+_34519790 | 0.00 |
ENSRNOT00000052360
|
Mgat1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr12_-_12330929 | 0.00 |
ENSRNOT00000001336
|
Bri3
|
brain protein I3 |
| chr4_+_52147641 | 0.00 |
ENSRNOT00000009458
|
Hyal4
|
hyaluronoglucosaminidase 4 |
| chr5_-_77408323 | 0.00 |
ENSRNOT00000046857
ENSRNOT00000046760 |
LOC259245
Mup4
|
alpha-2u globulin PGCL5 major urinary protein 4 |
| chr17_+_15852548 | 0.00 |
ENSRNOT00000022203
|
Card19
|
caspase recruitment domain family, member 19 |
| chr3_+_260653 | 0.00 |
ENSRNOT00000006710
|
AABR07051190.1
|
|
| chr8_-_109560747 | 0.00 |
ENSRNOT00000087334
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr7_-_124172979 | 0.00 |
ENSRNOT00000074960
|
Arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr7_+_97559841 | 0.00 |
ENSRNOT00000007326
|
Zhx2
|
zinc fingers and homeoboxes 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mecom
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.0 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |