Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
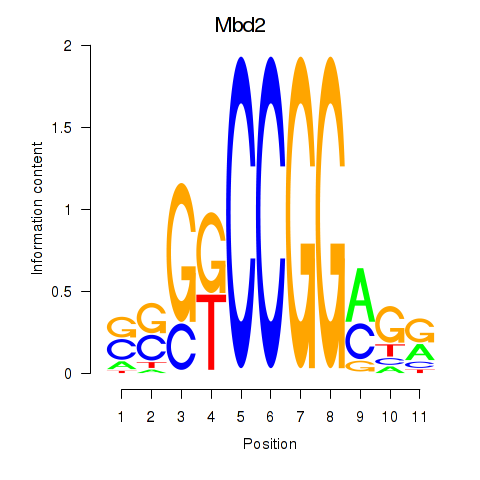
Results for Mbd2
Z-value: 0.57
Transcription factors associated with Mbd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mbd2
|
ENSRNOG00000011853 | methyl-CpG binding domain protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mbd2 | rn6_v1_chr18_+_65814026_65814026 | 0.50 | 3.9e-01 | Click! |
Activity profile of Mbd2 motif
Sorted Z-values of Mbd2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_78902063 | 0.35 |
ENSRNOT00000088469
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr10_+_85301875 | 0.34 |
ENSRNOT00000080935
|
Socs7
|
suppressor of cytokine signaling 7 |
| chr6_-_109162267 | 0.27 |
ENSRNOT00000077518
|
Nek9
|
NIMA-related kinase 9 |
| chr1_-_267245636 | 0.24 |
ENSRNOT00000082799
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr6_-_27190126 | 0.22 |
ENSRNOT00000068412
ENSRNOT00000013107 |
Kcnk3
|
potassium two pore domain channel subfamily K member 3 |
| chr3_-_164095878 | 0.22 |
ENSRNOT00000079414
|
B4galt5
|
beta-1,4-galactosyltransferase 5 |
| chr16_+_19051965 | 0.21 |
ENSRNOT00000016399
|
Slc35e1
|
solute carrier family 35, member E1 |
| chr19_-_53688597 | 0.20 |
ENSRNOT00000074448
|
Zcchc14
|
zinc finger CCHC-type containing 14 |
| chr15_+_108908607 | 0.20 |
ENSRNOT00000089455
|
Zic2
|
Zic family member 2 |
| chr8_+_117486083 | 0.19 |
ENSRNOT00000027552
ENSRNOT00000088705 |
Prkar2a
|
protein kinase cAMP-dependent type 2 regulatory subunit alpha |
| chr10_-_63952726 | 0.18 |
ENSRNOT00000090461
|
Doc2b
|
double C2 domain beta |
| chr10_+_62674561 | 0.16 |
ENSRNOT00000019946
ENSRNOT00000056110 |
Ankrd13b
|
ankyrin repeat domain 13B |
| chr10_-_90393317 | 0.16 |
ENSRNOT00000028563
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chrX_-_73778595 | 0.15 |
ENSRNOT00000076081
ENSRNOT00000075926 ENSRNOT00000003782 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr7_+_130498199 | 0.15 |
ENSRNOT00000092684
ENSRNOT00000092431 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr9_+_82571269 | 0.14 |
ENSRNOT00000026941
|
Speg
|
SPEG complex locus |
| chr17_-_10208360 | 0.13 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr19_+_23389375 | 0.13 |
ENSRNOT00000018629
|
Sall1
|
spalt-like transcription factor 1 |
| chr7_-_12085226 | 0.13 |
ENSRNOT00000088669
ENSRNOT00000079925 |
Onecut3
|
one cut domain, family member 3 |
| chr6_+_54488038 | 0.12 |
ENSRNOT00000084661
ENSRNOT00000005637 |
Snx13
|
sorting nexin 13 |
| chr8_+_48805684 | 0.12 |
ENSRNOT00000064041
|
Bcl9l
|
B-cell CLL/lymphoma 9-like |
| chr7_-_63045728 | 0.12 |
ENSRNOT00000039532
|
Lemd3
|
LEM domain containing 3 |
| chr6_-_126582034 | 0.12 |
ENSRNOT00000010656
ENSRNOT00000080829 |
Itpk1
|
inositol-tetrakisphosphate 1-kinase |
| chr5_-_74366817 | 0.11 |
ENSRNOT00000076705
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr8_+_122197027 | 0.11 |
ENSRNOT00000013050
|
Ubp1
|
upstream binding protein 1 (LBP-1a) |
| chr10_+_43067299 | 0.10 |
ENSRNOT00000003447
|
Galnt10
|
polypeptide N-acetylgalactosaminyltransferase 10 |
| chr1_-_164977633 | 0.10 |
ENSRNOT00000029629
|
Rnf169
|
ring finger protein 169 |
| chr2_-_24923128 | 0.10 |
ENSRNOT00000044087
|
Pde8b
|
phosphodiesterase 8B |
| chr2_-_115891097 | 0.10 |
ENSRNOT00000013191
|
Skil
|
SKI-like proto-oncogene |
| chr10_-_109267500 | 0.09 |
ENSRNOT00000005977
|
Cep131
|
centrosomal protein 131 |
| chr3_-_83306781 | 0.09 |
ENSRNOT00000014088
|
Ttc17
|
tetratricopeptide repeat domain 17 |
| chr7_-_140546908 | 0.09 |
ENSRNOT00000077502
|
Kmt2d
|
lysine methyltransferase 2D |
| chr3_+_93495106 | 0.09 |
ENSRNOT00000029922
ENSRNOT00000085760 |
Abtb2
|
ankyrin repeat and BTB domain containing 2 |
| chr12_-_47793534 | 0.09 |
ENSRNOT00000001588
|
Fam222a
|
family with sequence similarity 222, member A |
| chr7_+_25919867 | 0.09 |
ENSRNOT00000009625
ENSRNOT00000090153 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr10_-_82197848 | 0.09 |
ENSRNOT00000081307
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr10_+_14035149 | 0.09 |
ENSRNOT00000065827
|
Zfp598
|
zinc finger protein 598 |
| chr2_-_197935567 | 0.09 |
ENSRNOT00000085404
|
Rprd2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr7_+_23403891 | 0.08 |
ENSRNOT00000037918
|
Syn3
|
synapsin III |
| chr1_+_165724451 | 0.08 |
ENSRNOT00000025827
|
Fam168a
|
family with sequence similarity 168, member A |
| chr10_-_110182291 | 0.08 |
ENSRNOT00000015178
ENSRNOT00000054936 |
Csnk1d
|
casein kinase 1, delta |
| chr18_-_63394690 | 0.08 |
ENSRNOT00000090078
ENSRNOT00000029431 |
Cep76
|
centrosomal protein 76 |
| chr1_+_80087684 | 0.08 |
ENSRNOT00000041215
|
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr3_-_1924827 | 0.08 |
ENSRNOT00000006162
|
Cacna1b
|
calcium voltage-gated channel subunit alpha1 B |
| chr1_-_198460126 | 0.08 |
ENSRNOT00000082940
ENSRNOT00000086019 |
Maz
|
MYC associated zinc finger protein |
| chr2_+_205783252 | 0.08 |
ENSRNOT00000025633
ENSRNOT00000025600 |
Trim33
|
tripartite motif-containing 33 |
| chr6_+_137997335 | 0.08 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr15_-_4022223 | 0.07 |
ENSRNOT00000081997
|
Zswim8
|
zinc finger, SWIM-type containing 8 |
| chr7_-_141093924 | 0.07 |
ENSRNOT00000086276
|
Nckap5l
|
NCK-associated protein 5-like |
| chr6_-_46631983 | 0.07 |
ENSRNOT00000045963
|
Sox11
|
SRY box 11 |
| chr5_+_58393233 | 0.06 |
ENSRNOT00000000142
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr10_-_20818128 | 0.06 |
ENSRNOT00000011061
|
Wwc1
|
WW and C2 domain containing 1 |
| chr10_-_82197520 | 0.06 |
ENSRNOT00000092024
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr13_+_90532326 | 0.06 |
ENSRNOT00000008944
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr18_-_77322690 | 0.06 |
ENSRNOT00000058382
|
Nfatc1
|
nuclear factor of activated T-cells 1 |
| chr1_+_221644867 | 0.06 |
ENSRNOT00000054830
|
Ehd1
|
EH-domain containing 1 |
| chr3_+_170550314 | 0.06 |
ENSRNOT00000006991
|
Tfap2c
|
transcription factor AP-2 gamma |
| chr5_+_58393603 | 0.06 |
ENSRNOT00000080082
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr4_-_115157263 | 0.06 |
ENSRNOT00000015296
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr17_-_80320681 | 0.06 |
ENSRNOT00000023637
|
C1ql3
|
complement C1q like 3 |
| chr1_+_221612584 | 0.05 |
ENSRNOT00000090100
|
Atg2a
|
autophagy related 2A |
| chr2_+_198231291 | 0.05 |
ENSRNOT00000078288
ENSRNOT00000046739 |
Otud7b
|
OTU deubiquitinase 7B |
| chr19_+_56054212 | 0.05 |
ENSRNOT00000022324
|
Zfp276
|
zinc finger protein (C2H2 type) 276 |
| chr11_+_86512797 | 0.05 |
ENSRNOT00000051680
|
Gp1bb
|
glycoprotein Ib platelet beta subunit |
| chr3_+_147772165 | 0.05 |
ENSRNOT00000008713
|
Tbc1d20
|
TBC1 domain family, member 20 |
| chrX_-_15327684 | 0.05 |
ENSRNOT00000009794
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr17_-_75886523 | 0.05 |
ENSRNOT00000046266
|
Usp6nl
|
USP6 N-terminal like |
| chr5_+_148193710 | 0.05 |
ENSRNOT00000088568
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr1_-_221015929 | 0.05 |
ENSRNOT00000028137
|
Sipa1
|
signal-induced proliferation-associated 1 |
| chr1_+_251421596 | 0.05 |
ENSRNOT00000028143
|
Pten
|
phosphatase and tensin homolog |
| chrX_+_120859968 | 0.05 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr10_-_108196217 | 0.05 |
ENSRNOT00000075440
|
Cbx4
|
chromobox 4 |
| chr5_+_156876706 | 0.04 |
ENSRNOT00000021864
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr5_+_135735825 | 0.04 |
ENSRNOT00000068267
|
Zswim5
|
zinc finger, SWIM-type containing 5 |
| chr15_+_25933483 | 0.04 |
ENSRNOT00000018972
|
Ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr14_-_37871051 | 0.04 |
ENSRNOT00000003087
|
Slain2
|
SLAIN motif family, member 2 |
| chr2_+_43266739 | 0.04 |
ENSRNOT00000087031
|
Mier3
|
mesoderm induction early response 1, family member 3 |
| chr4_-_153028129 | 0.04 |
ENSRNOT00000074558
|
Cecr6
|
cat eye syndrome chromosome region, candidate 6 |
| chr1_-_127599257 | 0.04 |
ENSRNOT00000018436
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chr5_+_173183990 | 0.04 |
ENSRNOT00000068062
|
Tmem240
|
transmembrane protein 240 |
| chr8_-_103554399 | 0.04 |
ENSRNOT00000079965
ENSRNOT00000089833 |
LOC100909709
|
short transient receptor potential channel 1-like |
| chr10_+_91254058 | 0.04 |
ENSRNOT00000087218
ENSRNOT00000065373 |
Fmnl1
|
formin-like 1 |
| chr1_+_239398043 | 0.03 |
ENSRNOT00000087386
|
Tmem2
|
transmembrane protein 2 |
| chr6_-_102196138 | 0.03 |
ENSRNOT00000014132
|
Tmem229b
|
transmembrane protein 229B |
| chr5_+_144779681 | 0.03 |
ENSRNOT00000016493
|
LOC100294508
|
dyslexia susceptibility 2-like |
| chr10_+_82800704 | 0.03 |
ENSRNOT00000089497
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr14_+_83510278 | 0.03 |
ENSRNOT00000081161
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr2_-_206274079 | 0.03 |
ENSRNOT00000056079
|
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr4_-_163119391 | 0.03 |
ENSRNOT00000086605
|
Chtopl1
|
chromatin target of PRMT1-like 1 |
| chr4_+_32373641 | 0.03 |
ENSRNOT00000076086
|
Dlx6
|
distal-less homeobox 6 |
| chr5_+_136683592 | 0.03 |
ENSRNOT00000085527
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr8_-_111850075 | 0.03 |
ENSRNOT00000082097
|
Cdv3
|
CDV3 homolog |
| chr5_+_150459713 | 0.03 |
ENSRNOT00000081681
ENSRNOT00000074251 |
Taf12
|
TATA-box binding protein associated factor 12 |
| chr7_+_120125633 | 0.03 |
ENSRNOT00000012480
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr14_+_75852060 | 0.03 |
ENSRNOT00000075975
|
Hs3st1
|
heparan sulfate-glucosamine 3-sulfotransferase 1 |
| chr9_-_44786041 | 0.03 |
ENSRNOT00000067197
|
Rev1
|
REV1, DNA directed polymerase |
| chr7_+_137680530 | 0.03 |
ENSRNOT00000006970
|
Arid2
|
AT-rich interaction domain 2 |
| chr3_+_108795235 | 0.03 |
ENSRNOT00000007028
|
Spred1
|
sprouty-related, EVH1 domain containing 1 |
| chr10_-_35800120 | 0.02 |
ENSRNOT00000004361
|
Maml1
|
mastermind-like transcriptional coactivator 1 |
| chr14_+_15059387 | 0.02 |
ENSRNOT00000002843
|
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr19_-_56054142 | 0.02 |
ENSRNOT00000051945
|
Vps9d1
|
VPS9 domain containing 1 |
| chr8_-_68966108 | 0.02 |
ENSRNOT00000012155
|
Smad6
|
SMAD family member 6 |
| chr5_+_60250546 | 0.02 |
ENSRNOT00000017707
|
Zcchc7
|
zinc finger CCHC-type containing 7 |
| chr3_-_163748996 | 0.02 |
ENSRNOT00000010633
|
Stau1
|
staufen double-stranded RNA binding protein 1 |
| chr1_-_72727112 | 0.02 |
ENSRNOT00000031172
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr16_-_18860086 | 0.02 |
ENSRNOT00000074814
|
Sin3b
|
SIN3 transcription regulator family member B |
| chr2_-_210088949 | 0.02 |
ENSRNOT00000070994
|
Rbm15
|
RNA binding motif protein 15 |
| chr18_-_28495937 | 0.02 |
ENSRNOT00000027194
|
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr2_-_84437084 | 0.02 |
ENSRNOT00000077598
|
Ankrd33b
|
ankyrin repeat domain 33B |
| chrX_+_120860178 | 0.02 |
ENSRNOT00000088661
|
Wdr44
|
WD repeat domain 44 |
| chr10_-_95212111 | 0.02 |
ENSRNOT00000020795
|
Kpna2
|
karyopherin subunit alpha 2 |
| chr10_+_56605140 | 0.01 |
ENSRNOT00000024079
|
Phf23
|
PHD finger protein 23 |
| chr3_+_114747410 | 0.01 |
ENSRNOT00000043414
|
Spata5l1
|
spermatogenesis associated 5-like 1 |
| chr6_+_2569013 | 0.01 |
ENSRNOT00000008988
|
Atl2
|
atlastin GTPase 2 |
| chr1_-_265798167 | 0.01 |
ENSRNOT00000079483
|
Ldb1
|
LIM domain binding 1 |
| chr1_+_225140863 | 0.01 |
ENSRNOT00000027141
|
Mta2
|
metastasis associated 1 family, member 2 |
| chr2_+_218951141 | 0.01 |
ENSRNOT00000091001
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr1_+_206900617 | 0.01 |
ENSRNOT00000054897
|
Dock1
|
dedicator of cyto-kinesis 1 |
| chr7_-_70969905 | 0.01 |
ENSRNOT00000057745
|
Nab2
|
Ngfi-A binding protein 2 |
| chr2_-_52531083 | 0.01 |
ENSRNOT00000089303
ENSRNOT00000021608 |
Zfp131
|
zinc finger protein 131 |
| chr8_-_111850393 | 0.01 |
ENSRNOT00000044956
|
Cdv3
|
CDV3 homolog |
| chr17_+_72240802 | 0.01 |
ENSRNOT00000025993
ENSRNOT00000090453 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr3_-_176958880 | 0.01 |
ENSRNOT00000078661
|
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr10_-_84698886 | 0.01 |
ENSRNOT00000067542
|
Nfe2l1
|
nuclear factor, erythroid 2-like 1 |
| chr4_-_62663200 | 0.01 |
ENSRNOT00000040969
|
Cnot4
|
CCR4-NOT transcription complex, subunit 4 |
| chr8_-_48652064 | 0.01 |
ENSRNOT00000030745
ENSRNOT00000083490 |
C2cd2l
|
C2CD2-like |
| chr5_+_166533181 | 0.01 |
ENSRNOT00000045063
|
Clstn1
|
calsyntenin 1 |
| chr10_+_4957326 | 0.01 |
ENSRNOT00000003458
|
Socs1
|
suppressor of cytokine signaling 1 |
| chr2_-_218951030 | 0.01 |
ENSRNOT00000019172
|
Slc30a7
|
solute carrier family 30 member 7 |
| chr12_+_23473270 | 0.00 |
ENSRNOT00000001935
|
Sh2b2
|
SH2B adaptor protein 2 |
| chrX_-_15428518 | 0.00 |
ENSRNOT00000075774
|
LOC108348172
|
proSAAS |
| chr8_+_115069095 | 0.00 |
ENSRNOT00000014770
|
Dusp7
|
dual specificity phosphatase 7 |
| chr7_-_130408187 | 0.00 |
ENSRNOT00000015374
|
Chkb
|
choline kinase beta |
| chr15_+_83707735 | 0.00 |
ENSRNOT00000057843
|
Klf5
|
Kruppel-like factor 5 |
| chr8_-_49025917 | 0.00 |
ENSRNOT00000078816
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr4_+_57715946 | 0.00 |
ENSRNOT00000079059
ENSRNOT00000034429 |
Klhdc10
|
kelch domain containing 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mbd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1900095 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.2 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.2 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 0.1 | GO:0010045 | response to nickel cation(GO:0010045) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.0 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.0 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0051766 | inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |