Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
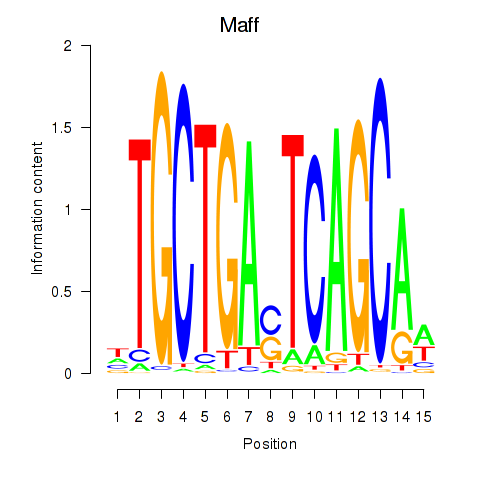
Results for Maff
Z-value: 0.31
Transcription factors associated with Maff
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Maff
|
ENSRNOG00000012886 | MAF bZIP transcription factor F |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Maff | rn6_v1_chr7_+_120580743_120580743 | 0.38 | 5.3e-01 | Click! |
Activity profile of Maff motif
Sorted Z-values of Maff motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_19714324 | 0.14 |
ENSRNOT00000072303
|
RGD1559588
|
similar to cell surface receptor FDFACT |
| chr2_-_14701903 | 0.09 |
ENSRNOT00000051895
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr8_+_128829680 | 0.09 |
ENSRNOT00000025279
|
Mobp
|
myelin-associated oligodendrocyte basic protein |
| chr14_+_34389991 | 0.08 |
ENSRNOT00000002953
|
Pdcl2
|
phosducin-like 2 |
| chr18_+_74006046 | 0.08 |
ENSRNOT00000086400
|
AABR07032622.1
|
|
| chr18_-_28454756 | 0.08 |
ENSRNOT00000040091
|
Spata24
|
spermatogenesis associated 24 |
| chr12_-_50404550 | 0.08 |
ENSRNOT00000073072
|
Crybb1
|
crystallin, beta B1 |
| chr7_-_29171783 | 0.07 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr13_+_52854031 | 0.06 |
ENSRNOT00000013738
ENSRNOT00000086004 |
Tmem9
|
transmembrane protein 9 |
| chr10_-_13168217 | 0.06 |
ENSRNOT00000087768
|
Elob
|
elongin B |
| chr20_+_6351458 | 0.06 |
ENSRNOT00000091731
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr2_-_198439454 | 0.06 |
ENSRNOT00000028780
|
Fcgr1a
|
Fc fragment of IgG receptor Ia |
| chr1_+_264507985 | 0.06 |
ENSRNOT00000085811
|
Pax2
|
paired box 2 |
| chrX_+_13441558 | 0.06 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr16_+_19881122 | 0.05 |
ENSRNOT00000060404
|
Dda1
|
DET1 and DDB1 associated 1 |
| chrX_-_123788898 | 0.05 |
ENSRNOT00000009123
|
Akap14
|
A-kinase anchoring protein 14 |
| chr1_-_175676699 | 0.05 |
ENSRNOT00000030474
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr1_-_225128740 | 0.05 |
ENSRNOT00000026897
|
Rom1
|
retinal outer segment membrane protein 1 |
| chr15_+_33596301 | 0.04 |
ENSRNOT00000022563
|
Il25
|
interleukin 25 |
| chr14_+_42402081 | 0.04 |
ENSRNOT00000092933
|
Bend4
|
BEN domain containing 4 |
| chr1_+_236580034 | 0.04 |
ENSRNOT00000074939
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr13_-_36101411 | 0.04 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr1_-_89483988 | 0.03 |
ENSRNOT00000028603
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr16_+_20027348 | 0.03 |
ENSRNOT00000034589
|
Fam129c
|
family with sequence similarity 129, member C |
| chr16_-_20800646 | 0.03 |
ENSRNOT00000083281
|
Comp
|
cartilage oligomeric matrix protein |
| chr4_-_157294047 | 0.03 |
ENSRNOT00000005601
|
Eno2
|
enolase 2 |
| chr14_-_33031282 | 0.03 |
ENSRNOT00000043244
|
LOC103693375
|
60S ribosomal protein L39 |
| chr9_-_95290931 | 0.03 |
ENSRNOT00000025300
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3 |
| chr9_+_20251521 | 0.03 |
ENSRNOT00000005535
|
LOC100911625
|
gamma-enolase-like |
| chr3_-_11410732 | 0.03 |
ENSRNOT00000034930
|
RGD1561113
|
similar to Hypothetical UPF0184 protein C9orf16 homolog |
| chr9_-_82154266 | 0.03 |
ENSRNOT00000024283
|
Cryba2
|
crystallin, beta A2 |
| chr6_-_86822094 | 0.03 |
ENSRNOT00000006531
|
Fkbp3
|
FK506 binding protein 3 |
| chr15_+_28028521 | 0.03 |
ENSRNOT00000089631
|
AC114343.1
|
|
| chr1_+_48273611 | 0.03 |
ENSRNOT00000022254
ENSRNOT00000022068 |
Slc22a1
|
solute carrier family 22 member 1 |
| chr18_+_55666027 | 0.02 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chr3_-_148407778 | 0.02 |
ENSRNOT00000011262
|
Foxs1
|
forkhead box S1 |
| chr1_-_85317968 | 0.02 |
ENSRNOT00000026891
|
Gmfg
|
glia maturation factor, gamma |
| chr7_+_51794173 | 0.02 |
ENSRNOT00000043774
|
Otogl
|
otogelin-like |
| chr10_-_74001895 | 0.02 |
ENSRNOT00000005658
|
Vmp1
|
vacuole membrane protein 1 |
| chrX_-_10772633 | 0.02 |
ENSRNOT00000035153
|
Ube2q2l
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr4_-_21920651 | 0.02 |
ENSRNOT00000066211
|
Tmem243
|
transmembrane protein 243 |
| chr8_-_109576353 | 0.02 |
ENSRNOT00000010320
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr3_-_122920684 | 0.02 |
ENSRNOT00000028818
|
Cpxm1
|
carboxypeptidase X (M14 family), member 1 |
| chr16_-_19349080 | 0.02 |
ENSRNOT00000038494
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr19_-_25288335 | 0.02 |
ENSRNOT00000050214
|
Nanos3
|
nanos C2HC-type zinc finger 3 |
| chr10_+_56187020 | 0.02 |
ENSRNOT00000046490
|
Tp53
|
tumor protein p53 |
| chr19_-_43848937 | 0.02 |
ENSRNOT00000044844
|
Ldhd
|
lactate dehydrogenase D |
| chr1_+_88955135 | 0.02 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chr10_-_63240332 | 0.02 |
ENSRNOT00000005125
|
Blmh
|
bleomycin hydrolase |
| chr7_+_91384187 | 0.01 |
ENSRNOT00000005828
|
Utp23
|
UTP23, small subunit processome component |
| chr5_-_40237591 | 0.01 |
ENSRNOT00000011393
|
Fut9
|
fucosyltransferase 9 |
| chr1_-_88826302 | 0.01 |
ENSRNOT00000028275
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr6_-_26281300 | 0.01 |
ENSRNOT00000078750
|
RGD1560110
|
similar to RIKEN cDNA 4930548H24 |
| chr6_-_49089855 | 0.01 |
ENSRNOT00000006526
|
Tpo
|
thyroid peroxidase |
| chr8_-_1450138 | 0.01 |
ENSRNOT00000008062
|
Aasdhppt
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr10_+_87808493 | 0.01 |
ENSRNOT00000065405
|
LOC680428
|
hypothetical protein LOC680428 |
| chr2_+_189582991 | 0.01 |
ENSRNOT00000022627
|
Rab13
|
RAB13, member RAS oncogene family |
| chr6_+_52702544 | 0.01 |
ENSRNOT00000014252
|
Efcab10
|
EF-hand calcium binding domain 10 |
| chr1_-_170073473 | 0.01 |
ENSRNOT00000085141
|
Olr196
|
olfactory receptor 196 |
| chr1_+_87009730 | 0.01 |
ENSRNOT00000027537
|
Ech1
|
enoyl-CoA hydratase 1 |
| chr7_+_121361415 | 0.01 |
ENSRNOT00000067904
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr10_-_43931094 | 0.01 |
ENSRNOT00000039778
|
Olr1415
|
olfactory receptor 1415 |
| chr13_-_79899479 | 0.01 |
ENSRNOT00000035815
|
RGD1309106
|
similar to hypothetical protein |
| chr6_+_102392828 | 0.01 |
ENSRNOT00000089162
|
Rdh12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis) |
| chr13_-_111765944 | 0.01 |
ENSRNOT00000073041
|
Syt14
|
synaptotagmin 14 |
| chr1_-_221448570 | 0.01 |
ENSRNOT00000075165
|
Zfpl1
|
zinc finger protein-like 1 |
| chr1_+_114046478 | 0.01 |
ENSRNOT00000032254
|
Siglech
|
sialic acid binding Ig-like lectin H |
| chr1_+_219390523 | 0.01 |
ENSRNOT00000054852
|
Gpr152
|
G protein-coupled receptor 152 |
| chr5_+_48224994 | 0.01 |
ENSRNOT00000068028
|
Rragd
|
Ras-related GTP binding D |
| chr1_+_275957698 | 0.01 |
ENSRNOT00000021041
|
Tectb
|
tectorin beta |
| chr10_+_74002151 | 0.01 |
ENSRNOT00000005661
|
Ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr1_-_62114672 | 0.01 |
ENSRNOT00000072607
|
AABR07001923.1
|
|
| chr19_-_49510901 | 0.01 |
ENSRNOT00000082929
ENSRNOT00000079969 |
LOC687399
|
hypothetical protein LOC687399 |
| chr1_-_73226777 | 0.01 |
ENSRNOT00000025080
|
Ncr1
|
natural cytotoxicity triggering receptor 1 |
| chrX_-_45806198 | 0.01 |
ENSRNOT00000066663
|
Prrg1
|
proline rich and Gla domain 1 |
| chr16_-_14382641 | 0.01 |
ENSRNOT00000018723
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr6_-_78817641 | 0.01 |
ENSRNOT00000072117
|
AABR07064392.1
|
|
| chr1_+_221448661 | 0.01 |
ENSRNOT00000072493
|
LOC100910252
|
sororin-like |
| chr5_+_127571114 | 0.00 |
ENSRNOT00000016557
|
Slc1a7
|
solute carrier family 1 member 7 |
| chr3_-_72289310 | 0.00 |
ENSRNOT00000038250
|
Rtn4rl2
|
reticulon 4 receptor-like 2 |
| chr9_+_47386626 | 0.00 |
ENSRNOT00000021270
|
Slc9a2
|
solute carrier family 9 member A2 |
| chr18_+_40883224 | 0.00 |
ENSRNOT00000039733
|
Lvrn
|
laeverin |
| chr5_+_127571312 | 0.00 |
ENSRNOT00000090817
ENSRNOT00000077808 ENSRNOT00000072263 |
Slc1a7
|
solute carrier family 1 member 7 |
| chr2_+_52397175 | 0.00 |
ENSRNOT00000082614
|
Ccl28
|
C-C motif chemokine ligand 28 |
| chr17_+_52938980 | 0.00 |
ENSRNOT00000081052
|
Gabpb1l
|
GA binding protein transcription factor, beta subunit 1-like |
| chr1_-_102013243 | 0.00 |
ENSRNOT00000042115
|
Abcc6
|
ATP binding cassette subfamily C member 6 |
| chr15_-_109394905 | 0.00 |
ENSRNOT00000019602
ENSRNOT00000078424 |
Tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr9_+_79659251 | 0.00 |
ENSRNOT00000021656
|
Xrcc5
|
X-ray repair cross complementing 5 |
| chr10_-_93675991 | 0.00 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr1_+_31700953 | 0.00 |
ENSRNOT00000020251
|
Exoc3
|
exocyst complex component 3 |
| chr2_-_188727670 | 0.00 |
ENSRNOT00000074920
|
Lenep
|
lens epithelial protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Maff
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.0 | 0.1 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |