Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
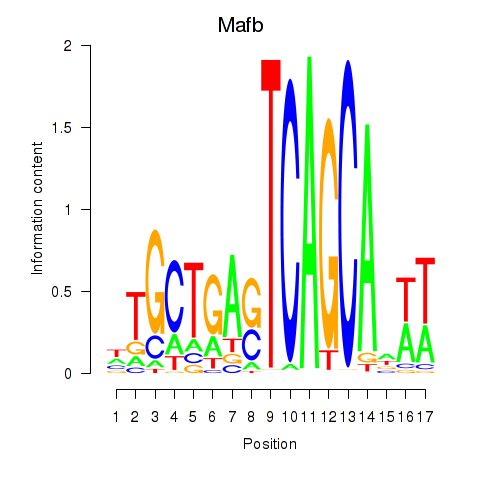
Results for Mafb
Z-value: 0.71
Transcription factors associated with Mafb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mafb
|
ENSRNOG00000016037 | MAF bZIP transcription factor B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mafb | rn6_v1_chr3_-_156340913_156340913 | -0.75 | 1.5e-01 | Click! |
Activity profile of Mafb motif
Sorted Z-values of Mafb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_50404550 | 0.19 |
ENSRNOT00000073072
|
Crybb1
|
crystallin, beta B1 |
| chr2_-_199479309 | 0.17 |
ENSRNOT00000042661
|
Rpl38
|
ribosomal protein L38 |
| chrX_+_13441558 | 0.17 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chrX_+_136460215 | 0.17 |
ENSRNOT00000093538
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr14_-_21709084 | 0.17 |
ENSRNOT00000087477
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chr6_+_86131242 | 0.16 |
ENSRNOT00000039337
|
Fau
|
FAU, ubiquitin like and ribosomal protein S30 fusion |
| chr5_+_16845631 | 0.16 |
ENSRNOT00000047889
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr1_-_103323476 | 0.16 |
ENSRNOT00000019051
|
Mrgprx3
|
MAS related GPR family member X3 |
| chr8_-_40883880 | 0.15 |
ENSRNOT00000075593
|
LOC102550797
|
disks large homolog 5-like |
| chr2_-_14701903 | 0.15 |
ENSRNOT00000051895
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr12_+_19714324 | 0.15 |
ENSRNOT00000072303
|
RGD1559588
|
similar to cell surface receptor FDFACT |
| chr5_-_38923095 | 0.14 |
ENSRNOT00000009146
|
AABR07047593.1
|
|
| chr10_-_19715832 | 0.14 |
ENSRNOT00000040278
|
RGD1564698
|
similar to ribosomal protein S10 |
| chr1_-_162713610 | 0.14 |
ENSRNOT00000018091
|
Aqp11
|
aquaporin 11 |
| chr8_+_98745310 | 0.14 |
ENSRNOT00000019938
|
Zic4
|
Zic family member 4 |
| chr9_-_73948583 | 0.14 |
ENSRNOT00000018097
|
Myl1
|
myosin, light chain 1 |
| chr10_+_49538588 | 0.14 |
ENSRNOT00000081415
|
Pmp22
|
peripheral myelin protein 22 |
| chrX_-_42329232 | 0.13 |
ENSRNOT00000045705
|
LOC100361934
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 |
| chr12_-_41224254 | 0.13 |
ENSRNOT00000085747
|
Oas1h
|
2 ' -5 ' oligoadenylate synthetase 1H |
| chr4_-_30338679 | 0.13 |
ENSRNOT00000012050
|
Pon3
|
paraoxonase 3 |
| chr1_-_81412251 | 0.13 |
ENSRNOT00000026946
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr11_+_82415995 | 0.13 |
ENSRNOT00000051772
|
AABR07034641.1
|
|
| chr10_-_13051154 | 0.13 |
ENSRNOT00000033457
|
AABR07029202.1
|
|
| chr9_+_45605552 | 0.13 |
ENSRNOT00000059587
|
NMS
|
neuromedin S |
| chr9_+_81656116 | 0.13 |
ENSRNOT00000083421
|
Slc11a1
|
solute carrier family 11 member 1 |
| chr18_+_56071478 | 0.13 |
ENSRNOT00000025344
ENSRNOT00000025354 |
Cd74
|
CD74 molecule |
| chr2_-_84531192 | 0.13 |
ENSRNOT00000065312
ENSRNOT00000090540 |
Ropn1l
|
rhophilin associated tail protein 1-like |
| chr1_+_264507985 | 0.13 |
ENSRNOT00000085811
|
Pax2
|
paired box 2 |
| chr16_-_79838212 | 0.13 |
ENSRNOT00000016756
|
Cln8
|
ceroid-lipofuscinosis, neuronal 8 |
| chr2_+_193572053 | 0.12 |
ENSRNOT00000056479
|
AABR07012329.1
|
|
| chr4_-_117531480 | 0.12 |
ENSRNOT00000021120
|
Nat8f5
|
N-acetyltransferase 8 (GCN5-related) family member 5 |
| chr5_+_137189473 | 0.12 |
ENSRNOT00000056815
|
Hyi
|
hydroxypyruvate isomerase |
| chr10_+_69423086 | 0.12 |
ENSRNOT00000000256
|
Ccl7
|
C-C motif chemokine ligand 7 |
| chr5_-_155252003 | 0.12 |
ENSRNOT00000017060
|
C1qb
|
complement C1q B chain |
| chr5_-_2803855 | 0.12 |
ENSRNOT00000009490
|
LOC297756
|
ribosomal protein S8-like |
| chr1_-_206394346 | 0.12 |
ENSRNOT00000038122
|
LOC100302465
|
hypothetical LOC100302465 |
| chr10_+_104952458 | 0.12 |
ENSRNOT00000074082
|
RGD1559482
|
similar to immunoglobulin superfamily, member 7 |
| chrX_-_40086870 | 0.12 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr5_+_159606647 | 0.12 |
ENSRNOT00000051693
|
Rps21-ps1
|
ribosomal protein S21, pseudogene 1 |
| chr15_+_38096994 | 0.12 |
ENSRNOT00000064635
|
Mrpl57
|
mitochondrial ribosomal protein L57 |
| chr2_-_198439454 | 0.12 |
ENSRNOT00000028780
|
Fcgr1a
|
Fc fragment of IgG receptor Ia |
| chr19_-_345698 | 0.12 |
ENSRNOT00000044037
|
LOC501297
|
hypothetical LOC501297 |
| chr14_-_70098021 | 0.12 |
ENSRNOT00000004912
|
Med28
|
mediator complex subunit 28 |
| chr3_-_10153554 | 0.12 |
ENSRNOT00000093246
|
Exosc2
|
exosome component 2 |
| chrX_+_29504349 | 0.12 |
ENSRNOT00000005981
|
Tceanc
|
transcription elongation factor A N-terminal and central domain containing |
| chr10_+_14122878 | 0.11 |
ENSRNOT00000052008
|
Hs3st6
|
heparan sulfate-glucosamine 3-sulfotransferase 6 |
| chr9_+_8399632 | 0.11 |
ENSRNOT00000092165
|
LOC100360856
|
hypothetical protein LOC100360856 |
| chr13_-_36101411 | 0.11 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr5_+_164923302 | 0.11 |
ENSRNOT00000042285
|
LOC108349682
|
60S ribosomal protein L35a |
| chr16_+_10250404 | 0.11 |
ENSRNOT00000087037
ENSRNOT00000081183 |
Gdf10
|
growth differentiation factor 10 |
| chr20_+_29951637 | 0.11 |
ENSRNOT00000074387
|
LOC100361018
|
rCG22048-like |
| chr10_-_14613878 | 0.11 |
ENSRNOT00000024072
|
Baiap3
|
BAI1-associated protein 3 |
| chr9_-_101388833 | 0.11 |
ENSRNOT00000071817
|
Chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chrM_+_7758 | 0.10 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr18_-_81682206 | 0.10 |
ENSRNOT00000058219
|
LOC100359752
|
hypothetical protein LOC100359752 |
| chr1_-_89084859 | 0.10 |
ENSRNOT00000032026
|
Cox6b1
|
cytochrome c oxidase subunit 6B1 |
| chr1_+_106896790 | 0.10 |
ENSRNOT00000042952
ENSRNOT00000064193 |
Ano5
|
anoctamin 5 |
| chr11_+_32450587 | 0.10 |
ENSRNOT00000061235
|
Smim11
|
small integral membrane protein 11 |
| chr10_-_46404642 | 0.10 |
ENSRNOT00000083698
|
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr10_-_104163634 | 0.10 |
ENSRNOT00000005198
|
Mif4gd
|
MIF4G domain containing |
| chrX_-_15761932 | 0.10 |
ENSRNOT00000015641
ENSRNOT00000091146 |
Foxp3
|
forkhead box P3 |
| chr14_-_33031282 | 0.10 |
ENSRNOT00000043244
|
LOC103693375
|
60S ribosomal protein L39 |
| chr7_+_11490852 | 0.10 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr7_-_29171783 | 0.10 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr1_-_175676699 | 0.09 |
ENSRNOT00000030474
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chrX_+_83926513 | 0.09 |
ENSRNOT00000035274
|
RGD1561958
|
similar to RIKEN cDNA 2010106E10 |
| chr17_+_9282675 | 0.09 |
ENSRNOT00000051702
|
H2afy
|
H2A histone family, member Y |
| chr11_+_53081025 | 0.09 |
ENSRNOT00000002700
|
Bbx
|
BBX, HMG-box containing |
| chr19_-_10212050 | 0.09 |
ENSRNOT00000082822
|
Tepp
|
testis, prostate and placenta expressed |
| chr17_-_2705123 | 0.09 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr11_+_64861768 | 0.09 |
ENSRNOT00000038439
|
Adprh
|
ADP-ribosylarginine hydrolase |
| chr13_-_79899479 | 0.09 |
ENSRNOT00000035815
|
RGD1309106
|
similar to hypothetical protein |
| chr18_-_15225427 | 0.09 |
ENSRNOT00000090853
|
Rnf125
|
ring finger protein 125 |
| chr2_+_198359754 | 0.09 |
ENSRNOT00000048582
|
Hist2h2ab
|
histone cluster 2 H2A family member b |
| chr6_-_86822094 | 0.09 |
ENSRNOT00000006531
|
Fkbp3
|
FK506 binding protein 3 |
| chr15_-_12129220 | 0.09 |
ENSRNOT00000029318
|
Olr1605
|
olfactory receptor 1605 |
| chr14_+_7949239 | 0.08 |
ENSRNOT00000044617
|
Ndufb4l1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4-like 1 |
| chr4_+_181103774 | 0.08 |
ENSRNOT00000084207
ENSRNOT00000055473 ENSRNOT00000077619 |
Arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr7_+_18700344 | 0.08 |
ENSRNOT00000009067
|
Cd320
|
CD320 molecule |
| chr4_+_3043231 | 0.08 |
ENSRNOT00000013732
|
Il6
|
interleukin 6 |
| chr18_+_74006046 | 0.08 |
ENSRNOT00000086400
|
AABR07032622.1
|
|
| chr2_+_195940715 | 0.08 |
ENSRNOT00000042985
|
LOC100363268
|
rCG31129-like |
| chr10_+_23914894 | 0.08 |
ENSRNOT00000071435
|
Ebf1
|
early B-cell factor 1 |
| chr7_+_63352046 | 0.08 |
ENSRNOT00000046453
|
RGD1561871
|
similar to ribosomal protein S10 |
| chr10_-_89374516 | 0.08 |
ENSRNOT00000028084
|
Vat1
|
vesicle amine transport 1 |
| chr15_+_28028521 | 0.08 |
ENSRNOT00000089631
|
AC114343.1
|
|
| chr2_-_189602143 | 0.08 |
ENSRNOT00000032062
|
Creb3l4
|
cAMP responsive element binding protein 3-like 4 |
| chr8_+_54925607 | 0.08 |
ENSRNOT00000059210
|
Plet1
|
placenta expressed transcript 1 |
| chr6_-_59531574 | 0.08 |
ENSRNOT00000042029
|
RGD1560073
|
similar to ribosomal protein S10 |
| chr10_+_56662561 | 0.08 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr11_+_67555658 | 0.08 |
ENSRNOT00000039075
|
Csta
|
cystatin A (stefin A) |
| chr17_+_15845931 | 0.08 |
ENSRNOT00000092083
|
Card19
|
caspase recruitment domain family, member 19 |
| chr9_+_81655629 | 0.08 |
ENSRNOT00000088679
ENSRNOT00000057472 |
Slc11a1
|
solute carrier family 11 member 1 |
| chr7_+_54980120 | 0.08 |
ENSRNOT00000005690
ENSRNOT00000005773 |
Kcnc2
|
potassium voltage-gated channel subfamily C member 2 |
| chr12_+_19890749 | 0.08 |
ENSRNOT00000074970
|
RGD1559588
|
similar to cell surface receptor FDFACT |
| chr13_+_52854031 | 0.08 |
ENSRNOT00000013738
ENSRNOT00000086004 |
Tmem9
|
transmembrane protein 9 |
| chr2_+_127845034 | 0.08 |
ENSRNOT00000044804
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr8_+_94821247 | 0.08 |
ENSRNOT00000081848
|
Mrap2
|
melanocortin 2 receptor accessory protein 2 |
| chr2_+_78103387 | 0.08 |
ENSRNOT00000063951
|
LOC103689968
|
protein FAM134B |
| chr6_-_21135880 | 0.08 |
ENSRNOT00000051239
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chr4_-_159192526 | 0.07 |
ENSRNOT00000026731
|
Kcna1
|
potassium voltage-gated channel subfamily A member 1 |
| chr1_+_128606770 | 0.07 |
ENSRNOT00000073355
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr15_+_30413488 | 0.07 |
ENSRNOT00000071781
|
AABR07017733.1
|
|
| chr10_-_87954055 | 0.07 |
ENSRNOT00000018048
|
Krt34
|
keratin 34 |
| chr2_-_206222248 | 0.07 |
ENSRNOT00000026075
|
Olfml3
|
olfactomedin-like 3 |
| chr17_-_10622226 | 0.07 |
ENSRNOT00000044559
|
Simc1
|
SUMO-interacting motifs containing 1 |
| chr1_+_238222521 | 0.07 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr20_-_4391402 | 0.07 |
ENSRNOT00000090518
ENSRNOT00000083489 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr7_-_18180705 | 0.07 |
ENSRNOT00000065954
|
Zfp494
|
zinc finger protein 494 |
| chr20_+_8486265 | 0.07 |
ENSRNOT00000072213
|
LOC100909911
|
40S ribosomal protein S20-like |
| chr9_+_100076958 | 0.07 |
ENSRNOT00000084297
|
Dusp28
|
dual specificity phosphatase 28 |
| chr14_-_22890958 | 0.07 |
ENSRNOT00000031916
|
LOC100911104
|
lymphocyte antigen 6B-like |
| chr3_-_137969658 | 0.07 |
ENSRNOT00000007727
|
Bfsp1
|
beaded filament structural protein 1 |
| chr9_+_65172194 | 0.07 |
ENSRNOT00000040493
|
AC128084.1
|
|
| chr16_+_20027348 | 0.07 |
ENSRNOT00000034589
|
Fam129c
|
family with sequence similarity 129, member C |
| chr10_+_47490153 | 0.07 |
ENSRNOT00000003182
|
Aldh3a1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr7_+_91832988 | 0.07 |
ENSRNOT00000006410
|
Slc30a8
|
solute carrier family 30 member 8 |
| chr19_+_25946979 | 0.07 |
ENSRNOT00000004027
|
Gadd45gip1
|
GADD45G interacting protein 1 |
| chr2_-_188413219 | 0.07 |
ENSRNOT00000065065
|
Fdps
|
farnesyl diphosphate synthase |
| chrX_+_131617798 | 0.07 |
ENSRNOT00000074384
|
Prr32
|
proline rich 32 |
| chr10_+_56662242 | 0.07 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr15_-_28092023 | 0.07 |
ENSRNOT00000090835
|
Eddm3b
|
epididymal protein 3B |
| chr5_-_134776247 | 0.07 |
ENSRNOT00000057055
|
Dmbx1
|
diencephalon/mesencephalon homeobox 1 |
| chr6_+_52702544 | 0.07 |
ENSRNOT00000014252
|
Efcab10
|
EF-hand calcium binding domain 10 |
| chr14_+_58877806 | 0.07 |
ENSRNOT00000051559
|
RGD1562755
|
similar to 60S ribosomal protein L23a |
| chr18_+_30023828 | 0.07 |
ENSRNOT00000079008
|
Pcdha4
|
protocadherin alpha 4 |
| chr10_-_82229140 | 0.07 |
ENSRNOT00000004400
|
Epn3
|
epsin 3 |
| chr13_+_85580828 | 0.07 |
ENSRNOT00000005611
|
Aldh9a1
|
aldehyde dehydrogenase 9 family, member A1 |
| chr13_+_48689962 | 0.07 |
ENSRNOT00000079707
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr5_-_9429859 | 0.07 |
ENSRNOT00000009462
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr7_-_123476238 | 0.07 |
ENSRNOT00000036917
|
Cenpm
|
centromere protein M |
| chr8_+_76426335 | 0.07 |
ENSRNOT00000085496
|
Gtf2a2
|
general transcription factor2A subunit 2 |
| chr1_-_85317968 | 0.06 |
ENSRNOT00000026891
|
Gmfg
|
glia maturation factor, gamma |
| chrM_+_7919 | 0.06 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr1_+_165189985 | 0.06 |
ENSRNOT00000048437
|
Kcne3
|
potassium voltage-gated channel subfamily E regulatory subunit 3 |
| chr18_-_48384645 | 0.06 |
ENSRNOT00000023485
|
Ppic
|
peptidylprolyl isomerase C |
| chr18_-_55859333 | 0.06 |
ENSRNOT00000025989
|
Myoz3
|
myozenin 3 |
| chr17_+_43661222 | 0.06 |
ENSRNOT00000022881
ENSRNOT00000022809 ENSRNOT00000022810 |
Hfe
|
hemochromatosis |
| chr1_-_77830399 | 0.06 |
ENSRNOT00000052231
|
LOC100360449
|
ribosomal protein L9-like |
| chr1_-_12952906 | 0.06 |
ENSRNOT00000078193
|
LOC100360362
|
hypothetical protein LOC100360362 |
| chr1_+_199720038 | 0.06 |
ENSRNOT00000027297
|
Ahsp
|
alpha hemoglobin stabilizing protein |
| chr1_-_89014189 | 0.06 |
ENSRNOT00000028413
|
Lin37
|
lin-37 DREAM MuvB core complex component |
| chr1_-_167698263 | 0.06 |
ENSRNOT00000093046
|
Trim21
|
tripartite motif-containing 21 |
| chr4_+_22082194 | 0.06 |
ENSRNOT00000091799
|
Crot
|
carnitine O-octanoyltransferase |
| chr9_+_8349033 | 0.06 |
ENSRNOT00000073775
|
LOC100360856
|
hypothetical protein LOC100360856 |
| chr9_-_120101098 | 0.06 |
ENSRNOT00000002917
|
Fam114a1l1
|
family with sequence similarity 114, member A1-like 1 |
| chr1_+_48433079 | 0.06 |
ENSRNOT00000037369
|
Slc22a3
|
solute carrier family 22 member 3 |
| chr1_-_91588609 | 0.06 |
ENSRNOT00000050931
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr6_+_36089433 | 0.06 |
ENSRNOT00000090438
ENSRNOT00000005731 |
Nt5c1b
|
5'-nucleotidase, cytosolic IB |
| chr1_+_156262031 | 0.06 |
ENSRNOT00000038279
|
Tmem126b
|
transmembrane protein 126B |
| chr7_+_91384187 | 0.06 |
ENSRNOT00000005828
|
Utp23
|
UTP23, small subunit processome component |
| chr3_-_104018861 | 0.06 |
ENSRNOT00000008387
|
Chrm5
|
cholinergic receptor, muscarinic 5 |
| chr13_-_48927483 | 0.06 |
ENSRNOT00000010976
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr1_+_221057713 | 0.06 |
ENSRNOT00000028197
|
Kcnk7
|
potassium two pore domain channel subfamily K member 7 |
| chr20_+_4966817 | 0.06 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr1_-_116153722 | 0.06 |
ENSRNOT00000041605
|
Fpr3
|
formyl peptide receptor 3 |
| chr2_+_195665454 | 0.06 |
ENSRNOT00000081335
|
Tdrkh
|
tudor and KH domain containing |
| chr8_+_91368111 | 0.05 |
ENSRNOT00000083151
|
Ttk
|
Ttk protein kinase |
| chr1_-_183763664 | 0.05 |
ENSRNOT00000044231
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr20_+_48503973 | 0.05 |
ENSRNOT00000064081
|
Cdc40
|
cell division cycle 40 |
| chrX_+_40363646 | 0.05 |
ENSRNOT00000010135
|
Sms
|
spermine synthase |
| chr7_-_96464049 | 0.05 |
ENSRNOT00000006517
|
Has2
|
hyaluronan synthase 2 |
| chr3_+_44025300 | 0.05 |
ENSRNOT00000006319
|
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr1_-_143392532 | 0.05 |
ENSRNOT00000026089
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr1_+_56982821 | 0.05 |
ENSRNOT00000059845
|
Ermard
|
ER membrane-associated RNA degradation |
| chr11_+_71533078 | 0.05 |
ENSRNOT00000002405
|
Slc51a
|
solute carrier family 51, alpha subunit |
| chr10_+_103266296 | 0.05 |
ENSRNOT00000038852
|
Dnai2
|
dynein, axonemal, intermediate chain 2 |
| chr5_-_139749050 | 0.05 |
ENSRNOT00000056651
|
Nfyc
|
nuclear transcription factor Y subunit gamma |
| chr9_-_14618013 | 0.05 |
ENSRNOT00000018351
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chrX_-_139476206 | 0.05 |
ENSRNOT00000049856
|
Ftl1l1
|
ferritin light chain 1-like 1 |
| chr6_-_75551625 | 0.05 |
ENSRNOT00000005825
|
Sptssa
|
serine palmitoyltransferase, small subunit A |
| chr5_+_140923914 | 0.05 |
ENSRNOT00000020929
|
Heyl
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr20_-_4392343 | 0.05 |
ENSRNOT00000080476
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr10_-_88152064 | 0.05 |
ENSRNOT00000019477
|
Krt16
|
keratin 16 |
| chr9_+_20279938 | 0.05 |
ENSRNOT00000075612
|
Grcc10
|
gene rich cluster, C10 gene |
| chr1_+_44446765 | 0.05 |
ENSRNOT00000030132
|
Cldn20
|
claudin 20 |
| chr4_+_162078905 | 0.05 |
ENSRNOT00000049620
|
LOC100910725
|
C-type lectin domain family 2 member D-related protein-like |
| chr7_-_117140168 | 0.05 |
ENSRNOT00000056481
ENSRNOT00000013353 |
Puf60
|
poly-U binding splicing factor 60 |
| chr10_-_88144468 | 0.05 |
ENSRNOT00000084619
|
Ka11
|
type I keratin KA11 |
| chr12_-_19167015 | 0.05 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chr10_+_83201311 | 0.05 |
ENSRNOT00000006179
|
Slc35b1
|
solute carrier family 35, member B1 |
| chr2_-_93641497 | 0.05 |
ENSRNOT00000013720
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr10_-_10807079 | 0.05 |
ENSRNOT00000087029
|
LOC360479
|
similar to hypothetical protein |
| chr17_-_14627937 | 0.05 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr3_+_159392193 | 0.05 |
ENSRNOT00000078101
ENSRNOT00000066260 |
Ift52
|
intraflagellar transport 52 |
| chr17_-_21484456 | 0.05 |
ENSRNOT00000038180
|
Sycp2l
|
synaptonemal complex protein 2-like |
| chr5_-_131860637 | 0.05 |
ENSRNOT00000064569
ENSRNOT00000080242 |
Slc5a9
|
solute carrier family 5 member 9 |
| chr1_+_64480929 | 0.05 |
ENSRNOT00000087016
|
Olr1l
|
olfactory receptor 1-like |
| chr11_-_25078740 | 0.05 |
ENSRNOT00000002109
|
Cyyr1
|
cysteine and tyrosine rich 1 |
| chr8_-_71052529 | 0.05 |
ENSRNOT00000033512
|
Ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chrX_+_15098904 | 0.05 |
ENSRNOT00000007367
ENSRNOT00000087033 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr6_-_60124274 | 0.05 |
ENSRNOT00000059823
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr10_+_4953879 | 0.05 |
ENSRNOT00000003455
|
Tnp2
|
transition protein 2 |
| chr3_-_103745236 | 0.05 |
ENSRNOT00000006876
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr12_+_24148567 | 0.05 |
ENSRNOT00000077378
ENSRNOT00000045157 |
Ccl24
|
C-C motif chemokine ligand 24 |
| chr2_+_124150224 | 0.05 |
ENSRNOT00000078650
|
Spata5
|
spermatogenesis associated 5 |
| chr10_-_34333305 | 0.05 |
ENSRNOT00000071365
|
Olr1384
|
olfactory receptor gene Olr1384 |
| chr2_-_148838441 | 0.05 |
ENSRNOT00000018294
|
Erich6
|
glutamate-rich 6 |
| chr4_+_145413230 | 0.05 |
ENSRNOT00000056508
|
Il17re
|
interleukin 17 receptor E |
| chr10_-_104263071 | 0.05 |
ENSRNOT00000005347
|
Grb2
|
growth factor receptor bound protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mafb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.0 | 0.1 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.1 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.0 | 0.1 | GO:0071038 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0002465 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:1904815 | negative regulation of histone phosphorylation(GO:0033128) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0002585 | positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.2 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:1905132 | metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0019046 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.0 | 0.0 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.0 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.0 | GO:0019541 | propionate metabolic process(GO:0019541) |
| 0.0 | 0.1 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.0 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.0 | GO:2001226 | negative regulation of inorganic anion transmembrane transport(GO:1903796) negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.0 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.0 | GO:1903165 | response to polycyclic arene(GO:1903165) |
| 0.0 | 0.0 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.0 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.0 | GO:0032301 | recombination nodule(GO:0005713) MutSalpha complex(GO:0032301) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.0 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.0 | GO:0005688 | U6 snRNP(GO:0005688) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0016414 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.0 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.0 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |