Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
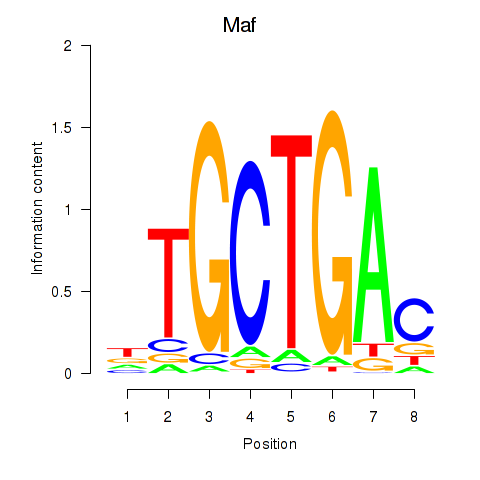
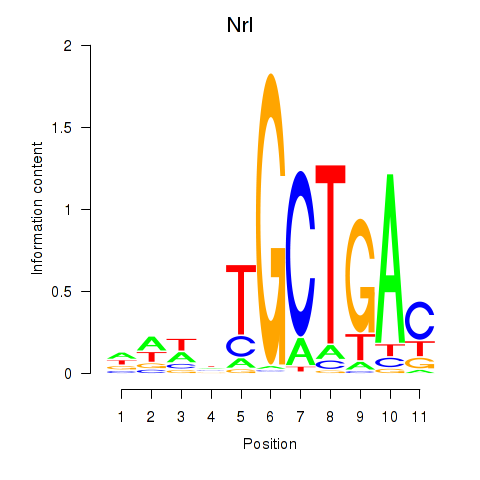
Results for Maf_Nrl
Z-value: 0.35
Transcription factors associated with Maf_Nrl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Maf
|
ENSRNOG00000012428 | MAF bZIP transcription factor |
|
Nrl
|
ENSRNOG00000018528 | neural retina leucine zipper |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Maf | rn6_v1_chr19_-_48196748_48196748 | -0.82 | 9.0e-02 | Click! |
| Nrl | rn6_v1_chr15_-_34198921_34198921 | 0.07 | 9.1e-01 | Click! |
Activity profile of Maf_Nrl motif
Sorted Z-values of Maf_Nrl motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_73948583 | 0.24 |
ENSRNOT00000018097
|
Myl1
|
myosin, light chain 1 |
| chr5_-_153924896 | 0.24 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr2_-_113616766 | 0.13 |
ENSRNOT00000016858
ENSRNOT00000074723 |
Tmem212
|
transmembrane protein 212 |
| chr15_-_32888095 | 0.12 |
ENSRNOT00000012233
|
Dad1
|
defender against cell death 1 |
| chr20_-_5163247 | 0.11 |
ENSRNOT00000001135
|
Aif1
|
allograft inflammatory factor 1 |
| chrX_+_128897181 | 0.11 |
ENSRNOT00000008444
|
Sh2d1a
|
SH2 domain containing 1A |
| chr2_-_28799266 | 0.10 |
ENSRNOT00000089293
|
Tmem171
|
transmembrane protein 171 |
| chrX_-_32153794 | 0.09 |
ENSRNOT00000005348
|
Tmem27
|
transmembrane protein 27 |
| chr1_+_276659542 | 0.08 |
ENSRNOT00000019681
|
Tcf7l2
|
transcription factor 7 like 2 |
| chr9_+_45605552 | 0.08 |
ENSRNOT00000059587
|
NMS
|
neuromedin S |
| chr12_+_50407843 | 0.08 |
ENSRNOT00000073763
|
Cryba4
|
crystallin, beta A4 |
| chr17_-_389967 | 0.07 |
ENSRNOT00000023865
|
Fbp2
|
fructose-bisphosphatase 2 |
| chr6_+_10533151 | 0.07 |
ENSRNOT00000020822
|
Rhoq
|
ras homolog family member Q |
| chr3_+_65815080 | 0.07 |
ENSRNOT00000006429
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr10_+_71202456 | 0.06 |
ENSRNOT00000076893
|
Hnf1b
|
HNF1 homeobox B |
| chr20_+_10438444 | 0.06 |
ENSRNOT00000071248
ENSRNOT00000075545 |
Cryaa
|
crystallin, alpha A |
| chr10_-_6870011 | 0.06 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
| chr3_+_112242270 | 0.06 |
ENSRNOT00000080533
ENSRNOT00000082876 |
Capn3
|
calpain 3 |
| chr6_+_73345392 | 0.06 |
ENSRNOT00000088378
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr1_+_32199810 | 0.06 |
ENSRNOT00000036714
|
Slc6a19
|
solute carrier family 6 member 19 |
| chr10_+_55924938 | 0.05 |
ENSRNOT00000087003
ENSRNOT00000057079 |
Trappc1
|
trafficking protein particle complex 1 |
| chr11_-_28527890 | 0.05 |
ENSRNOT00000002138
|
Cldn8
|
claudin 8 |
| chr4_+_70828894 | 0.05 |
ENSRNOT00000064892
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr5_+_124300477 | 0.05 |
ENSRNOT00000010100
|
C8b
|
complement C8 beta chain |
| chr4_-_67610710 | 0.05 |
ENSRNOT00000039067
|
Mrps33
|
mitochondrial ribosomal protein S33 |
| chr9_-_73958480 | 0.05 |
ENSRNOT00000017838
|
Myl1
|
myosin, light chain 1 |
| chr16_+_83358116 | 0.05 |
ENSRNOT00000031109
|
Rab20
|
RAB20, member RAS oncogene family |
| chr3_-_173839433 | 0.05 |
ENSRNOT00000075547
|
AC127963.1
|
|
| chr8_+_94693290 | 0.04 |
ENSRNOT00000013908
|
Cyb5r4
|
cytochrome b5 reductase 4 |
| chr5_+_128881062 | 0.04 |
ENSRNOT00000057216
|
Calr4
|
calreticulin 4 |
| chr6_+_136358057 | 0.04 |
ENSRNOT00000092741
|
Klc1
|
kinesin light chain 1 |
| chr1_-_175676699 | 0.04 |
ENSRNOT00000030474
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr16_+_26906716 | 0.04 |
ENSRNOT00000064297
|
Cpe
|
carboxypeptidase E |
| chr20_+_3744395 | 0.04 |
ENSRNOT00000086065
|
Tcf19
|
transcription factor 19 |
| chr17_-_16695126 | 0.04 |
ENSRNOT00000021550
|
Id4
|
inhibitor of DNA binding 4, HLH protein |
| chr3_-_45210474 | 0.04 |
ENSRNOT00000091777
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr7_-_96464049 | 0.04 |
ENSRNOT00000006517
|
Has2
|
hyaluronan synthase 2 |
| chrX_+_35869538 | 0.04 |
ENSRNOT00000058947
|
Ppef1
|
protein phosphatase with EF-hand domain 1 |
| chr20_+_34258791 | 0.04 |
ENSRNOT00000000468
|
Slc35f1
|
solute carrier family 35, member F1 |
| chr3_+_57770948 | 0.04 |
ENSRNOT00000038429
|
AC107446.2
|
|
| chr9_-_16934714 | 0.04 |
ENSRNOT00000061503
|
Crip3
|
cysteine-rich protein 3 |
| chr14_+_33010300 | 0.04 |
ENSRNOT00000002809
|
Igfbp7
|
insulin-like growth factor binding protein 7 |
| chr4_-_178168690 | 0.03 |
ENSRNOT00000020729
|
Sox5
|
SRY box 5 |
| chrX_-_23139694 | 0.03 |
ENSRNOT00000033656
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr14_-_8432195 | 0.03 |
ENSRNOT00000089800
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr11_+_17538063 | 0.03 |
ENSRNOT00000031889
ENSRNOT00000090878 |
Chodl
|
chondrolectin |
| chr10_-_88152064 | 0.03 |
ENSRNOT00000019477
|
Krt16
|
keratin 16 |
| chrX_-_138435391 | 0.03 |
ENSRNOT00000043258
|
Mbnl3
|
muscleblind-like splicing regulator 3 |
| chr4_+_176994129 | 0.03 |
ENSRNOT00000018734
|
Cmas
|
cytidine monophosphate N-acetylneuraminic acid synthetase |
| chr12_+_19680712 | 0.03 |
ENSRNOT00000081310
|
AABR07035561.2
|
|
| chr10_+_97647111 | 0.03 |
ENSRNOT00000055062
|
Gna13
|
G protein subunit alpha 13 |
| chr13_-_42885440 | 0.03 |
ENSRNOT00000038020
|
Nckap5
|
NCK-associated protein 5 |
| chr4_-_97784842 | 0.03 |
ENSRNOT00000007698
|
Gadd45a
|
growth arrest and DNA-damage-inducible, alpha |
| chr8_+_69121682 | 0.03 |
ENSRNOT00000013461
|
Rpl4
|
ribosomal protein L4 |
| chr10_+_90985653 | 0.03 |
ENSRNOT00000093364
|
Ccdc103
|
coiled-coil domain containing 103 |
| chr3_-_8659102 | 0.03 |
ENSRNOT00000050908
|
Zdhhc12
|
zinc finger, DHHC-type containing 12 |
| chr10_-_20622600 | 0.03 |
ENSRNOT00000020554
|
Fbll1
|
fibrillarin-like 1 |
| chrX_+_110818716 | 0.03 |
ENSRNOT00000086308
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr10_-_88144468 | 0.03 |
ENSRNOT00000084619
|
Ka11
|
type I keratin KA11 |
| chr9_+_100076958 | 0.03 |
ENSRNOT00000084297
|
Dusp28
|
dual specificity phosphatase 28 |
| chr20_-_5469966 | 0.02 |
ENSRNOT00000092362
|
Tapbp
|
TAP binding protein |
| chr13_+_27032048 | 0.02 |
ENSRNOT00000031789
|
Serpinb13
|
serpin family B member 13 |
| chr6_+_146394456 | 0.02 |
ENSRNOT00000007683
|
RGD1562420
|
similar to hypothetical protein |
| chr6_-_7421456 | 0.02 |
ENSRNOT00000006725
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr6_+_27975849 | 0.02 |
ENSRNOT00000060810
|
Dtnb
|
dystrobrevin, beta |
| chr5_+_136014017 | 0.02 |
ENSRNOT00000025346
|
Best4
|
bestrophin 4 |
| chr5_+_76150840 | 0.02 |
ENSRNOT00000020702
|
Dnajc25
|
DnaJ heat shock protein family (Hsp40) member C25 |
| chr12_-_19254527 | 0.02 |
ENSRNOT00000089349
|
AABR07035541.3
|
|
| chrX_-_135250519 | 0.02 |
ENSRNOT00000044487
|
Elf4
|
E74 like ETS transcription factor 4 |
| chr8_-_68312909 | 0.02 |
ENSRNOT00000066106
|
RGD1309779
|
similar to ENSANGP00000021391 |
| chr1_-_101514547 | 0.02 |
ENSRNOT00000079633
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr1_+_22364551 | 0.02 |
ENSRNOT00000043157
|
LOC108348200
|
trace amine-associated receptor 8a |
| chr1_-_141533908 | 0.02 |
ENSRNOT00000020117
|
Mesp1
|
mesoderm posterior bHLH transcription factor 1 |
| chr3_-_5693147 | 0.02 |
ENSRNOT00000008660
|
Fam163b
|
family with sequence similarity 163, member B |
| chr10_+_53818818 | 0.02 |
ENSRNOT00000057260
|
Myh8
|
myosin heavy chain 8 |
| chr11_+_27208564 | 0.02 |
ENSRNOT00000002158
|
Map3k7cl
|
MAP3K7 C-terminal like |
| chr1_+_169562247 | 0.02 |
ENSRNOT00000023124
|
Olr153
|
olfactory receptor 153 |
| chr5_+_28395296 | 0.02 |
ENSRNOT00000009375
|
Tmem55a
|
transmembrane protein 55A |
| chr18_-_26211445 | 0.02 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chrX_+_131381134 | 0.02 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr3_+_91191837 | 0.02 |
ENSRNOT00000006097
|
Rag2
|
recombination activating 2 |
| chr3_+_37545238 | 0.02 |
ENSRNOT00000070792
|
Tnfaip6
|
TNF alpha induced protein 6 |
| chr1_+_264507985 | 0.02 |
ENSRNOT00000085811
|
Pax2
|
paired box 2 |
| chr1_+_85003280 | 0.02 |
ENSRNOT00000057122
|
Fcgbp
|
Fc fragment of IgG binding protein |
| chr19_+_10167797 | 0.02 |
ENSRNOT00000085440
|
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr10_-_109774639 | 0.02 |
ENSRNOT00000054955
|
Alyref
|
Aly/REF export factor |
| chr16_-_47163898 | 0.02 |
ENSRNOT00000074490
|
Ccdc110
|
coiled-coil domain containing 110 |
| chr19_+_27404712 | 0.02 |
ENSRNOT00000023657
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_+_48718329 | 0.02 |
ENSRNOT00000089763
|
Slc37a4
|
solute carrier family 37 member 4 |
| chr9_+_81566074 | 0.02 |
ENSRNOT00000074131
ENSRNOT00000046229 ENSRNOT00000090383 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr1_-_240601744 | 0.02 |
ENSRNOT00000024093
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr20_+_295250 | 0.02 |
ENSRNOT00000000955
|
Clic2
|
chloride intracellular channel 2 |
| chr5_+_171297850 | 0.02 |
ENSRNOT00000034284
|
Lrrc47
|
leucine rich repeat containing 47 |
| chr1_+_102915191 | 0.02 |
ENSRNOT00000017851
|
Ldhc
|
lactate dehydrogenase C |
| chr4_+_6931495 | 0.02 |
ENSRNOT00000079770
|
Wdr86
|
WD repeat domain 86 |
| chr4_-_113886994 | 0.02 |
ENSRNOT00000037333
|
Htra2
|
HtrA serine peptidase 2 |
| chr14_-_2371883 | 0.02 |
ENSRNOT00000000077
|
Pde6b
|
phosphodiesterase 6B |
| chr4_-_56656980 | 0.02 |
ENSRNOT00000009278
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr12_+_22165486 | 0.01 |
ENSRNOT00000001890
|
Mospd3
|
motile sperm domain containing 3 |
| chr4_+_7282948 | 0.01 |
ENSRNOT00000011052
|
Cdk5
|
cyclin-dependent kinase 5 |
| chr19_-_37675077 | 0.01 |
ENSRNOT00000031033
|
Enkd1
|
enkurin domain containing 1 |
| chr16_+_8823872 | 0.01 |
ENSRNOT00000027186
|
Drgx
|
dorsal root ganglia homeobox |
| chr5_-_12172009 | 0.01 |
ENSRNOT00000061903
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr14_-_81426496 | 0.01 |
ENSRNOT00000018073
|
Add1
|
adducin 1 |
| chr9_+_42620006 | 0.01 |
ENSRNOT00000019966
|
Hs6st1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr18_-_57031459 | 0.01 |
ENSRNOT00000026583
|
Pcyox1l
|
prenylcysteine oxidase 1 like |
| chr15_+_2766710 | 0.01 |
ENSRNOT00000017483
|
Dupd1
|
dual specificity phosphatase and pro isomerase domain containing 1 |
| chr3_-_7498555 | 0.01 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr18_-_57515834 | 0.01 |
ENSRNOT00000026098
|
Adrb2
|
adrenoceptor beta 2 |
| chr1_-_157494008 | 0.01 |
ENSRNOT00000013552
|
Pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr15_-_3544685 | 0.01 |
ENSRNOT00000015179
ENSRNOT00000085126 |
Vcl
|
vinculin |
| chr3_-_133894569 | 0.01 |
ENSRNOT00000084221
|
AABR07053968.1
|
|
| chr4_-_40385349 | 0.01 |
ENSRNOT00000039005
|
Gpr85
|
G protein-coupled receptor 85 |
| chr13_-_53137253 | 0.01 |
ENSRNOT00000011368
|
Gpr25
|
G protein-coupled receptor 25 |
| chr15_-_27819376 | 0.01 |
ENSRNOT00000067400
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr3_+_120728688 | 0.01 |
ENSRNOT00000022596
|
Bcl2l11
|
BCL2 like 11 |
| chr2_+_18392142 | 0.01 |
ENSRNOT00000043196
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr13_+_52413241 | 0.01 |
ENSRNOT00000035157
|
AABR07020999.1
|
|
| chr20_+_37876650 | 0.01 |
ENSRNOT00000001054
|
Gja1
|
gap junction protein, alpha 1 |
| chr2_+_148722231 | 0.01 |
ENSRNOT00000018022
|
Eif2a
|
eukaryotic translation initiation factor 2A |
| chr2_-_138833933 | 0.01 |
ENSRNOT00000013343
|
Pcdh18
|
protocadherin 18 |
| chr9_-_19372673 | 0.01 |
ENSRNOT00000073667
ENSRNOT00000079517 |
Clic5
|
chloride intracellular channel 5 |
| chr10_-_98469799 | 0.01 |
ENSRNOT00000087502
ENSRNOT00000088646 |
Abca9
|
ATP binding cassette subfamily A member 9 |
| chr11_-_84037938 | 0.01 |
ENSRNOT00000002327
|
Abcf3
|
ATP binding cassette subfamily F member 3 |
| chr5_-_117612123 | 0.01 |
ENSRNOT00000065112
|
Dock7
|
dedicator of cytokinesis 7 |
| chr6_-_114488880 | 0.01 |
ENSRNOT00000087560
|
AC118957.1
|
|
| chr1_+_164225934 | 0.01 |
ENSRNOT00000034389
|
Map6
|
microtubule-associated protein 6 |
| chr17_+_11683862 | 0.01 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr4_+_88584242 | 0.01 |
ENSRNOT00000008973
|
Pyurf
|
PIGY upstream reading frame |
| chr19_+_52077501 | 0.01 |
ENSRNOT00000079240
|
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr2_+_11658568 | 0.01 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr18_+_29386809 | 0.01 |
ENSRNOT00000082079
ENSRNOT00000024825 |
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr3_-_110249627 | 0.01 |
ENSRNOT00000009866
|
Srp14
|
signal recognition particle 14 |
| chr5_+_156810991 | 0.01 |
ENSRNOT00000055904
|
AABR07050222.1
|
|
| chr1_-_8885967 | 0.01 |
ENSRNOT00000016102
|
Gje1
|
gap junction protein, epsilon 1 |
| chr10_+_65161152 | 0.01 |
ENSRNOT00000011608
|
Cryba1
|
crystallin, beta A1 |
| chr5_-_147584038 | 0.01 |
ENSRNOT00000010983
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chr11_-_33003021 | 0.01 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr9_+_18564927 | 0.01 |
ENSRNOT00000061014
|
Runx2
|
runt-related transcription factor 2 |
| chr18_-_63488027 | 0.01 |
ENSRNOT00000023763
|
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr10_-_63240332 | 0.00 |
ENSRNOT00000005125
|
Blmh
|
bleomycin hydrolase |
| chr3_-_10144388 | 0.00 |
ENSRNOT00000042495
|
Abl1
|
ABL proto-oncogene 1, non-receptor tyrosine kinase |
| chr1_+_238222521 | 0.00 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr14_+_83560541 | 0.00 |
ENSRNOT00000057738
ENSRNOT00000085228 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr6_+_132510757 | 0.00 |
ENSRNOT00000080230
|
Evl
|
Enah/Vasp-like |
| chr2_+_127845034 | 0.00 |
ENSRNOT00000044804
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr20_-_3822754 | 0.00 |
ENSRNOT00000000541
ENSRNOT00000077357 |
Slc39a7
|
solute carrier family 39 member 7 |
| chr2_-_148722263 | 0.00 |
ENSRNOT00000017868
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr9_-_99659425 | 0.00 |
ENSRNOT00000051686
|
Olr1343
|
olfactory receptor 1343 |
| chr11_-_87924816 | 0.00 |
ENSRNOT00000031819
|
Serpind1
|
serpin family D member 1 |
| chr18_+_40596166 | 0.00 |
ENSRNOT00000087984
|
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr1_-_101514974 | 0.00 |
ENSRNOT00000044788
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr1_-_214844858 | 0.00 |
ENSRNOT00000046344
|
Tollip
|
toll interacting protein |
| chr7_-_23948202 | 0.00 |
ENSRNOT00000044252
|
Ascl4
|
achaete-scute family bHLH transcription factor 4 |
| chr7_+_28412198 | 0.00 |
ENSRNOT00000081822
ENSRNOT00000038780 ENSRNOT00000005995 |
Igf1
|
insulin-like growth factor 1 |
| chr1_+_256955652 | 0.00 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
| chr4_-_82141385 | 0.00 |
ENSRNOT00000008447
|
Hoxa3
|
homeobox A3 |
| chr5_-_171327450 | 0.00 |
ENSRNOT00000088710
|
Ccdc27
|
coiled-coil domain containing 27 |
| chr15_+_83707735 | 0.00 |
ENSRNOT00000057843
|
Klf5
|
Kruppel-like factor 5 |
| chr11_+_27101227 | 0.00 |
ENSRNOT00000002173
|
Usp16
|
ubiquitin specific peptidase 16 |
| chr3_-_110140756 | 0.00 |
ENSRNOT00000007882
|
Gpr176
|
G protein-coupled receptor 176 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Maf_Nrl
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.0 | 0.1 | GO:0048619 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0061017 | regulation of pronephros size(GO:0035565) pronephric nephron tubule development(GO:0039020) hepatoblast differentiation(GO:0061017) |
| 0.0 | 0.1 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.2 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |