Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
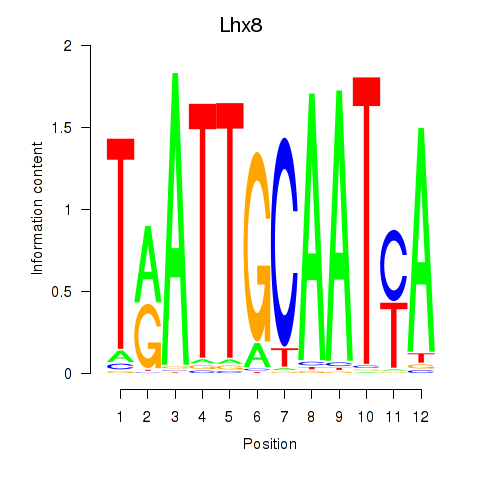
Results for Lhx8
Z-value: 0.43
Transcription factors associated with Lhx8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx8
|
ENSRNOG00000028348 | LIM homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx8 | rn6_v1_chr2_-_260596777_260596777 | -0.47 | 4.3e-01 | Click! |
Activity profile of Lhx8 motif
Sorted Z-values of Lhx8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_48891130 | 0.26 |
ENSRNOT00000083884
|
AC135026.1
|
|
| chr2_-_147819335 | 0.14 |
ENSRNOT00000057909
|
Ankub1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr1_+_64740487 | 0.14 |
ENSRNOT00000081213
|
LOC103691005
|
zinc finger protein 679-like |
| chr15_-_44442875 | 0.13 |
ENSRNOT00000017939
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr5_+_6373583 | 0.13 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr7_-_82059247 | 0.11 |
ENSRNOT00000087177
|
Rspo2
|
R-spondin 2 |
| chr1_-_49844547 | 0.11 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr7_-_40316532 | 0.10 |
ENSRNOT00000083347
|
RGD1307947
|
similar to RIKEN cDNA C430008C19 |
| chr2_-_170460754 | 0.10 |
ENSRNOT00000013009
|
Slitrk3
|
SLIT and NTRK-like family, member 3 |
| chr10_+_65552897 | 0.09 |
ENSRNOT00000056217
|
Spag5
|
sperm associated antigen 5 |
| chr7_-_129919946 | 0.09 |
ENSRNOT00000079976
|
AABR07058658.1
|
|
| chrM_+_9870 | 0.09 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr9_-_38495126 | 0.09 |
ENSRNOT00000016933
ENSRNOT00000090385 |
Rab23
|
RAB23, member RAS oncogene family |
| chr6_-_105160470 | 0.09 |
ENSRNOT00000008367
|
Adam4
|
a disintegrin and metalloprotease domain 4 |
| chr11_-_70499200 | 0.09 |
ENSRNOT00000002439
|
Slc12a8
|
solute carrier family 12, member 8 |
| chr16_+_18648866 | 0.09 |
ENSRNOT00000014862
|
Dydc1
|
DPY30 domain containing 1 |
| chr15_-_45947030 | 0.08 |
ENSRNOT00000035888
|
Ints6
|
integrator complex subunit 6 |
| chr16_-_49820235 | 0.08 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr2_-_86475096 | 0.07 |
ENSRNOT00000075247
ENSRNOT00000075062 |
LOC100909688
|
zinc finger protein 43-like |
| chr1_-_71373605 | 0.07 |
ENSRNOT00000034854
|
Galp
|
galanin-like peptide |
| chr4_-_159192526 | 0.07 |
ENSRNOT00000026731
|
Kcna1
|
potassium voltage-gated channel subfamily A member 1 |
| chr11_+_38727048 | 0.07 |
ENSRNOT00000081537
|
LOC103690343
|
zinc finger protein 260-like |
| chr2_+_235738416 | 0.07 |
ENSRNOT00000074209
|
Etnppl
|
ethanolamine-phosphate phospho-lyase |
| chr16_-_3765917 | 0.07 |
ENSRNOT00000088284
|
Duxbl1
|
double homeobox B-like 1 |
| chr4_-_150506557 | 0.07 |
ENSRNOT00000076927
|
Zfp248
|
zinc finger protein 248 |
| chr3_+_48106099 | 0.06 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr6_-_76608864 | 0.06 |
ENSRNOT00000010824
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr17_+_78915604 | 0.06 |
ENSRNOT00000057855
|
Rpp38
|
ribonuclease P/MRP 38 subunit |
| chr1_+_42169501 | 0.06 |
ENSRNOT00000025477
ENSRNOT00000092791 |
Vip
|
vasoactive intestinal peptide |
| chr5_-_164648328 | 0.06 |
ENSRNOT00000090811
|
Zfp933
|
zinc finger protein 933 |
| chr12_-_5773036 | 0.06 |
ENSRNOT00000041365
|
Fry
|
FRY microtubule binding protein |
| chr16_-_54450426 | 0.06 |
ENSRNOT00000014623
|
Pdgfrl
|
platelet-derived growth factor receptor-like |
| chr2_+_211546560 | 0.06 |
ENSRNOT00000033443
|
Aknad1
|
AKNA domain containing 1 |
| chr1_-_282492912 | 0.06 |
ENSRNOT00000047883
|
AABR07007130.1
|
|
| chr1_+_221710670 | 0.06 |
ENSRNOT00000064798
|
Map4k2
|
mitogen activated protein kinase kinase kinase kinase 2 |
| chr4_+_70252366 | 0.06 |
ENSRNOT00000073039
|
Chl1
|
cell adhesion molecule L1-like |
| chrM_+_8599 | 0.05 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr9_-_65442257 | 0.05 |
ENSRNOT00000037660
|
Fam126b
|
family with sequence similarity 126, member B |
| chr14_+_34354503 | 0.05 |
ENSRNOT00000002941
|
Nmu
|
neuromedin U |
| chr8_-_48582353 | 0.05 |
ENSRNOT00000011582
|
Pdzd3
|
PDZ domain containing 3 |
| chr4_-_150506406 | 0.05 |
ENSRNOT00000076307
|
Zfp248
|
zinc finger protein 248 |
| chr2_-_30634243 | 0.05 |
ENSRNOT00000077537
|
Marveld2
|
MARVEL domain containing 2 |
| chr6_+_9790422 | 0.05 |
ENSRNOT00000020959
|
Prkce
|
protein kinase C, epsilon |
| chrM_+_9451 | 0.05 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr5_-_168123030 | 0.05 |
ENSRNOT00000092747
|
Per3
|
period circadian clock 3 |
| chr20_+_4043741 | 0.05 |
ENSRNOT00000000525
|
RT1-Bb
|
RT1 class II, locus Bb |
| chr4_+_123760743 | 0.05 |
ENSRNOT00000013498
|
Ccdc174
|
coiled-coil domain containing 174 |
| chr8_-_48597867 | 0.05 |
ENSRNOT00000077958
|
Nlrx1
|
NLR family member X1 |
| chr18_-_1828606 | 0.05 |
ENSRNOT00000033312
|
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr5_-_16140896 | 0.05 |
ENSRNOT00000029503
|
Xkr4
|
XK related 4 |
| chr7_+_132378273 | 0.05 |
ENSRNOT00000010990
|
LOC690142
|
hypothetical protein LOC690142 |
| chr7_-_15301382 | 0.05 |
ENSRNOT00000081512
|
Zfp763
|
zinc finger protein 763 |
| chr17_+_34665872 | 0.04 |
ENSRNOT00000092101
ENSRNOT00000085704 |
Exoc2
|
exocyst complex component 2 |
| chr3_+_51687809 | 0.04 |
ENSRNOT00000087242
|
Scn2a
|
sodium voltage-gated channel alpha subunit 2 |
| chr11_-_71136673 | 0.04 |
ENSRNOT00000042240
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr3_-_2584523 | 0.04 |
ENSRNOT00000017373
|
Uap1l1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 like 1 |
| chr9_-_81772851 | 0.04 |
ENSRNOT00000032719
|
Usp37
|
ubiquitin specific peptidase 37 |
| chr2_-_22105710 | 0.04 |
ENSRNOT00000017705
|
Zfyve16
|
zinc finger FYVE-type containing 16 |
| chr1_-_73399377 | 0.04 |
ENSRNOT00000038898
|
Lilrb4
|
leukocyte immunoglobulin like receptor B4 |
| chr2_+_72006099 | 0.04 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr7_+_29909120 | 0.04 |
ENSRNOT00000049362
|
AABR07056556.1
|
|
| chr7_+_2831004 | 0.04 |
ENSRNOT00000029778
|
Rnf41
|
ring finger protein 41 |
| chr15_-_93868292 | 0.04 |
ENSRNOT00000093546
ENSRNOT00000014583 |
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr2_+_200076054 | 0.04 |
ENSRNOT00000025327
|
Sec22b
|
SEC22 homolog B, vesicle trafficking protein |
| chr1_+_218466289 | 0.04 |
ENSRNOT00000017948
|
Mrgprf
|
MAS related GPR family member F |
| chr10_-_52290657 | 0.04 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr9_+_65110330 | 0.03 |
ENSRNOT00000033068
ENSRNOT00000092868 |
Aox4
|
aldehyde oxidase 4 |
| chr12_+_12649861 | 0.03 |
ENSRNOT00000092402
|
Ccz1b
|
CCZ1 homolog B, vacuolar protein trafficking and biogenesis associated |
| chr1_-_67094567 | 0.03 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr12_+_16899025 | 0.03 |
ENSRNOT00000001716
|
Psmg3
|
proteasome assembly chaperone 3 |
| chr6_-_99870024 | 0.03 |
ENSRNOT00000010043
|
Rab15
|
RAB15, member RAS oncogene family |
| chr1_-_263885169 | 0.03 |
ENSRNOT00000030782
|
Chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr1_-_169456098 | 0.03 |
ENSRNOT00000030827
|
Trim30c
|
tripartite motif-containing 30C |
| chr1_+_22364551 | 0.03 |
ENSRNOT00000043157
|
LOC108348200
|
trace amine-associated receptor 8a |
| chr1_+_204959174 | 0.03 |
ENSRNOT00000023257
|
Zranb1
|
zinc finger RANBP2-type containing 1 |
| chr1_-_22404002 | 0.03 |
ENSRNOT00000044098
|
Taar8a
|
trace amine-associated receptor 8a |
| chr13_-_47154292 | 0.03 |
ENSRNOT00000005284
|
Cd55
|
CD55 molecule, decay accelerating factor for complement |
| chr1_-_247476827 | 0.03 |
ENSRNOT00000021298
|
Insl6
|
insulin-like 6 |
| chr2_-_181531978 | 0.03 |
ENSRNOT00000072029
|
Npy2r
|
neuropeptide Y receptor Y2 |
| chr8_+_13862651 | 0.03 |
ENSRNOT00000014605
|
Taf1d
|
TATA-box binding protein associated factor, RNA polymerase I subunit D |
| chr3_-_105470475 | 0.03 |
ENSRNOT00000011078
|
Gjd2
|
gap junction protein, delta 2 |
| chr4_+_69386698 | 0.03 |
ENSRNOT00000091655
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr1_-_62114672 | 0.03 |
ENSRNOT00000072607
|
AABR07001923.1
|
|
| chr7_+_116671948 | 0.02 |
ENSRNOT00000077773
ENSRNOT00000029711 |
Gli4
|
GLI family zinc finger 4 |
| chr11_-_87977368 | 0.02 |
ENSRNOT00000002546
|
Tmem191c
|
transmembrane protein 191C |
| chr2_-_38110567 | 0.02 |
ENSRNOT00000072212
|
Ipo11
|
importin 11 |
| chr11_+_74834050 | 0.02 |
ENSRNOT00000002333
|
Atp13a4
|
ATPase 13A4 |
| chrX_-_915953 | 0.02 |
ENSRNOT00000075264
|
Spaca5
|
sperm acrosome associated 5 |
| chr16_+_32457521 | 0.02 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chrX_+_71540895 | 0.02 |
ENSRNOT00000004692
ENSRNOT00000082967 |
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr6_+_43234526 | 0.02 |
ENSRNOT00000086808
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr6_+_136004214 | 0.02 |
ENSRNOT00000013695
|
NEWGENE_619861
|
eukaryotic translation initiation factor 5 |
| chr5_+_144581427 | 0.02 |
ENSRNOT00000015227
|
Clspn
|
claspin |
| chrX_-_13601069 | 0.02 |
ENSRNOT00000004686
|
Otc
|
ornithine carbamoyltransferase |
| chr7_-_130408187 | 0.02 |
ENSRNOT00000015374
|
Chkb
|
choline kinase beta |
| chr1_-_167884690 | 0.02 |
ENSRNOT00000091372
|
Olr61
|
olfactory receptor 61 |
| chr9_+_79659251 | 0.02 |
ENSRNOT00000021656
|
Xrcc5
|
X-ray repair cross complementing 5 |
| chr12_-_38274036 | 0.02 |
ENSRNOT00000063990
|
Kntc1
|
kinetochore associated 1 |
| chr7_-_15852930 | 0.02 |
ENSRNOT00000009270
|
LOC691422
|
similar to zinc finger protein 101 |
| chr2_-_258997138 | 0.02 |
ENSRNOT00000045020
ENSRNOT00000042256 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr2_-_18531210 | 0.02 |
ENSRNOT00000088313
|
Vcan
|
versican |
| chr4_-_71227872 | 0.02 |
ENSRNOT00000050392
|
Tcaf2
|
TRPM8 channel-associated factor 2 |
| chr16_+_35934970 | 0.02 |
ENSRNOT00000084707
ENSRNOT00000016474 |
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chrM_+_7919 | 0.02 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr2_-_57935334 | 0.02 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr7_+_130308532 | 0.02 |
ENSRNOT00000011941
|
Miox
|
myo-inositol oxygenase |
| chr2_+_225570721 | 0.02 |
ENSRNOT00000017076
|
Arhgap29
|
Rho GTPase activating protein 29 |
| chr1_-_55895636 | 0.02 |
ENSRNOT00000029537
|
Vom2r9
|
vomeronasal 2 receptor, 9 |
| chr3_-_173944396 | 0.02 |
ENSRNOT00000079019
|
Sycp2
|
synaptonemal complex protein 2 |
| chrX_-_29825439 | 0.02 |
ENSRNOT00000048155
|
Gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr4_+_2593876 | 0.02 |
ENSRNOT00000092992
|
Ube3c
|
ubiquitin protein ligase E3C |
| chr12_-_12782139 | 0.02 |
ENSRNOT00000001392
ENSRNOT00000079836 |
Eif2ak1
|
eukaryotic translation initiation factor 2 alpha kinase 1 |
| chr7_+_121361415 | 0.02 |
ENSRNOT00000067904
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr13_+_82355471 | 0.02 |
ENSRNOT00000030677
|
Sele
|
selectin E |
| chr1_+_152072665 | 0.02 |
ENSRNOT00000022538
|
Rab38
|
RAB38, member RAS oncogene family |
| chr5_+_113592919 | 0.02 |
ENSRNOT00000011336
|
Ift74
|
intraflagellar transport 74 |
| chr13_+_82355886 | 0.02 |
ENSRNOT00000076757
|
Sele
|
selectin E |
| chr18_+_27047382 | 0.02 |
ENSRNOT00000027691
ENSRNOT00000090264 |
Apc
|
APC, WNT signaling pathway regulator |
| chr10_+_90230711 | 0.02 |
ENSRNOT00000055185
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr16_-_23156962 | 0.02 |
ENSRNOT00000018543
|
Sh2d4a
|
SH2 domain containing 4A |
| chr3_-_38090526 | 0.01 |
ENSRNOT00000059430
|
Cacnb4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr9_+_16702460 | 0.01 |
ENSRNOT00000061432
|
Ptk7
|
protein tyrosine kinase 7 |
| chr3_+_17889972 | 0.01 |
ENSRNOT00000073021
|
AABR07051611.1
|
|
| chr18_+_86299463 | 0.01 |
ENSRNOT00000058152
|
Cd226
|
CD226 molecule |
| chr6_+_28515025 | 0.01 |
ENSRNOT00000088033
ENSRNOT00000005317 ENSRNOT00000081141 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr8_+_52127632 | 0.01 |
ENSRNOT00000079797
|
Cadm1
|
cell adhesion molecule 1 |
| chr12_-_10335499 | 0.01 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chr1_-_204087001 | 0.01 |
ENSRNOT00000051853
|
Cpxm2
|
carboxypeptidase X (M14 family), member 2 |
| chr1_-_167893061 | 0.01 |
ENSRNOT00000049401
|
Olr60
|
olfactory receptor 60 |
| chr10_-_64341066 | 0.01 |
ENSRNOT00000082825
ENSRNOT00000010010 |
Vps53
|
VPS53 GARP complex subunit |
| chr16_+_8211420 | 0.01 |
ENSRNOT00000079609
|
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr18_-_36579403 | 0.01 |
ENSRNOT00000025284
|
Lars
|
leucyl-tRNA synthetase |
| chr1_-_261371508 | 0.01 |
ENSRNOT00000019978
|
Avpi1
|
arginine vasopressin-induced 1 |
| chrX_-_2657155 | 0.01 |
ENSRNOT00000005630
|
Chst7
|
carbohydrate sulfotransferase 7 |
| chr4_-_51598771 | 0.01 |
ENSRNOT00000008325
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr10_-_13996565 | 0.01 |
ENSRNOT00000016221
ENSRNOT00000052138 |
Tsc2
|
tuberous sclerosis 2 |
| chr2_+_60180215 | 0.01 |
ENSRNOT00000084624
|
Prlr
|
prolactin receptor |
| chr18_+_63203063 | 0.01 |
ENSRNOT00000024144
|
Prelid3a
|
PRELI domain containing 3A |
| chr10_-_45812641 | 0.01 |
ENSRNOT00000038951
|
Zfp867
|
zinc finger protein 867 |
| chr4_-_130659697 | 0.01 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr14_-_87701884 | 0.01 |
ENSRNOT00000079338
|
Mospd1
|
motile sperm domain containing 1 |
| chr3_-_9936352 | 0.01 |
ENSRNOT00000011224
ENSRNOT00000042798 |
Fnbp1
|
formin binding protein 1 |
| chr13_+_82072497 | 0.01 |
ENSRNOT00000063810
ENSRNOT00000085135 |
Kifap3
|
kinesin-associated protein 3 |
| chr3_-_82757204 | 0.01 |
ENSRNOT00000012214
|
Accs
|
1-aminocyclopropane-1-carboxylate synthase homolog |
| chr6_+_95323579 | 0.01 |
ENSRNOT00000007369
|
Pcnx4
|
pecanex homolog 4 (Drosophila) |
| chr1_+_1771710 | 0.01 |
ENSRNOT00000080138
ENSRNOT00000073528 |
Nup43
|
nucleoporin 43 |
| chrX_-_56765893 | 0.01 |
ENSRNOT00000076283
|
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr2_-_198834925 | 0.01 |
ENSRNOT00000089831
|
Nudt17
|
nudix hydrolase 17 |
| chr4_+_160020472 | 0.01 |
ENSRNOT00000078802
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr14_+_99529284 | 0.01 |
ENSRNOT00000006897
ENSRNOT00000067134 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chrX_+_31054394 | 0.01 |
ENSRNOT00000037439
|
LOC688526
|
similar to ribonucleic acid binding protein S1 |
| chr10_+_10875983 | 0.01 |
ENSRNOT00000052265
|
LOC679823
|
similar to 60S ribosomal protein L37a |
| chr20_-_27782586 | 0.01 |
ENSRNOT00000001093
|
Dse
|
dermatan sulfate epimerase |
| chr15_-_47338976 | 0.01 |
ENSRNOT00000016562
|
RGD1565212
|
similar to 4930578I06Rik protein |
| chr13_+_103300932 | 0.01 |
ENSRNOT00000085214
ENSRNOT00000038264 |
Eprs
|
glutamyl-prolyl-tRNA synthetase |
| chr9_-_63641400 | 0.01 |
ENSRNOT00000087684
|
Satb2
|
SATB homeobox 2 |
| chr1_-_89094530 | 0.01 |
ENSRNOT00000031873
|
Etv2
|
ets variant 2 |
| chr7_+_40316639 | 0.01 |
ENSRNOT00000080150
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr11_-_70207361 | 0.01 |
ENSRNOT00000002444
|
Muc13
|
mucin 13, cell surface associated |
| chr17_-_45746753 | 0.01 |
ENSRNOT00000077475
|
RGD1564243
|
similar to brain Zn-finger protein |
| chr2_+_62150493 | 0.00 |
ENSRNOT00000080241
|
Zfr
|
zinc finger RNA binding protein |
| chr3_+_16846412 | 0.00 |
ENSRNOT00000074266
|
AABR07051551.1
|
|
| chr18_+_30424814 | 0.00 |
ENSRNOT00000073425
|
Pcdhb7
|
protocadherin beta 7 |
| chrM_+_10160 | 0.00 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr5_+_138470069 | 0.00 |
ENSRNOT00000076343
ENSRNOT00000064073 |
Zmynd12
|
zinc finger, MYND-type containing 12 |
| chr1_-_147077568 | 0.00 |
ENSRNOT00000074040
|
LOC100911002
|
synaptonemal complex protein 3-like |
| chr1_+_190229528 | 0.00 |
ENSRNOT00000088121
|
Abca16
|
ATP-binding cassette, subfamily A (ABC1), member 16 |
| chr10_+_106065712 | 0.00 |
ENSRNOT00000003690
|
Sec14l1
|
SEC14-like lipid binding 1 |
| chr7_-_114590119 | 0.00 |
ENSRNOT00000079599
|
Ptk2
|
protein tyrosine kinase 2 |
| chr11_-_79703736 | 0.00 |
ENSRNOT00000044279
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr8_-_87419564 | 0.00 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chr4_-_168517177 | 0.00 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.0 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0010034 | response to acetate(GO:0010034) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |