Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
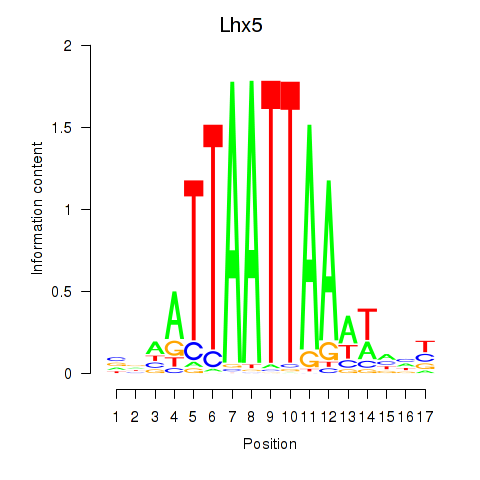
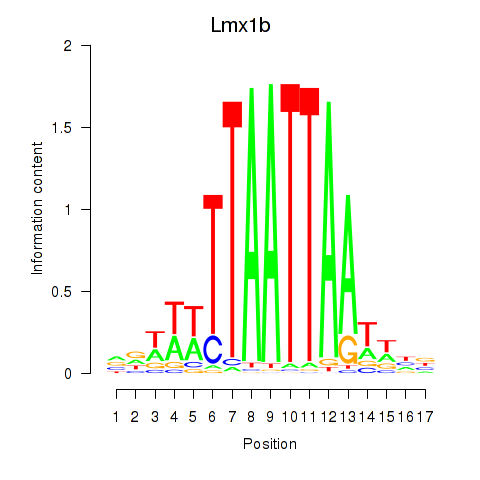
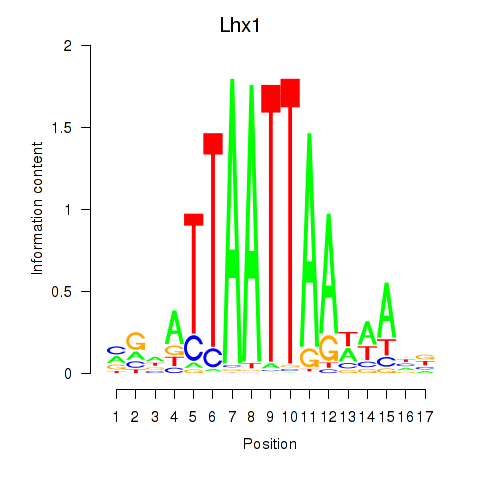
Results for Lhx5_Lmx1b_Lhx1
Z-value: 0.57
Transcription factors associated with Lhx5_Lmx1b_Lhx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx5
|
ENSRNOG00000001392 | LIM homeobox 5 |
|
Lmx1b
|
ENSRNOG00000017019 | LIM homeobox transcription factor 1 beta |
|
Lhx1
|
ENSRNOG00000002812 | LIM homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx1 | rn6_v1_chr10_-_71849293_71849293 | -0.50 | 3.9e-01 | Click! |
| Lhx5 | rn6_v1_chr12_-_41671437_41671437 | -0.46 | 4.3e-01 | Click! |
| Lmx1b | rn6_v1_chr3_-_12686869_12686869 | -0.10 | 8.8e-01 | Click! |
Activity profile of Lhx5_Lmx1b_Lhx1 motif
Sorted Z-values of Lhx5_Lmx1b_Lhx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_74679858 | 0.29 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr12_-_46493203 | 0.25 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr2_-_35870578 | 0.21 |
ENSRNOT00000066069
|
Rnf180
|
ring finger protein 180 |
| chr12_+_25435567 | 0.21 |
ENSRNOT00000092982
|
Gtf2i
|
general transcription factor II I |
| chr16_+_46731403 | 0.21 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr13_+_91054974 | 0.20 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr2_-_34452895 | 0.20 |
ENSRNOT00000079385
|
AABR07007905.2
|
|
| chr1_-_190370499 | 0.20 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr4_-_18396035 | 0.20 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr2_+_190003223 | 0.18 |
ENSRNOT00000015712
|
S100a5
|
S100 calcium binding protein A5 |
| chr2_+_127525285 | 0.18 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr8_-_39551700 | 0.17 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr20_-_13994794 | 0.17 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr16_+_84465656 | 0.16 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr6_+_49825469 | 0.16 |
ENSRNOT00000006921
|
Fam150b
|
family with sequence similarity 150, member B |
| chr15_-_93765498 | 0.15 |
ENSRNOT00000093297
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr17_+_9596957 | 0.14 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr3_+_14889510 | 0.14 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr1_+_101603222 | 0.14 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr17_-_18592750 | 0.13 |
ENSRNOT00000065742
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr14_+_113867209 | 0.13 |
ENSRNOT00000067591
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr20_+_42966140 | 0.13 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr6_-_21112734 | 0.12 |
ENSRNOT00000079819
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chr2_-_178521038 | 0.12 |
ENSRNOT00000033107
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr7_-_104801045 | 0.12 |
ENSRNOT00000079524
|
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr13_-_52413681 | 0.12 |
ENSRNOT00000037100
ENSRNOT00000011801 |
Nav1
|
neuron navigator 1 |
| chr2_+_86996497 | 0.11 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr18_-_55891710 | 0.11 |
ENSRNOT00000064686
|
Synpo
|
synaptopodin |
| chr13_-_76049363 | 0.11 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr18_+_30890869 | 0.11 |
ENSRNOT00000060466
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chrX_-_138118696 | 0.10 |
ENSRNOT00000084130
|
Frmd7
|
FERM domain containing 7 |
| chr2_+_86996798 | 0.10 |
ENSRNOT00000070821
|
Zfp455
|
zinc finger protein 455 |
| chr2_-_96509424 | 0.10 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr17_-_2278613 | 0.10 |
ENSRNOT00000046525
|
Cntnap3b
|
contactin associated protein-like 3B |
| chr20_+_3176107 | 0.10 |
ENSRNOT00000001036
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr1_+_100577056 | 0.10 |
ENSRNOT00000026992
|
Napsa
|
napsin A aspartic peptidase |
| chr7_-_104800536 | 0.10 |
ENSRNOT00000085650
|
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr9_-_30844199 | 0.09 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr2_-_33025271 | 0.09 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_-_39460844 | 0.09 |
ENSRNOT00000048875
|
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr1_-_88162583 | 0.09 |
ENSRNOT00000087411
|
Catsperg
|
cation channel sperm associated auxiliary subunit gamma |
| chr1_-_228014924 | 0.09 |
ENSRNOT00000042091
|
Oosp2
|
oocyte secreted protein 2 |
| chr10_+_61685645 | 0.08 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr8_-_126390801 | 0.08 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chr4_-_55011415 | 0.08 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr11_-_62451149 | 0.08 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr2_+_71753816 | 0.08 |
ENSRNOT00000025809
|
AABR07008724.1
|
|
| chr19_+_25043680 | 0.08 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr7_+_125649688 | 0.08 |
ENSRNOT00000017906
|
Phf21b
|
PHD finger protein 21B |
| chr9_+_73378057 | 0.08 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr3_-_45169118 | 0.08 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr18_-_43945273 | 0.08 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr2_-_38110567 | 0.07 |
ENSRNOT00000072212
|
Ipo11
|
importin 11 |
| chr20_+_3155652 | 0.07 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr13_+_57243877 | 0.07 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr10_+_69423086 | 0.07 |
ENSRNOT00000000256
|
Ccl7
|
C-C motif chemokine ligand 7 |
| chrX_+_71540895 | 0.07 |
ENSRNOT00000004692
ENSRNOT00000082967 |
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr11_-_43022565 | 0.07 |
ENSRNOT00000002285
|
Riox2
|
ribosomal oxygenase 2 |
| chr1_-_66237501 | 0.07 |
ENSRNOT00000073006
|
Zfp606
|
zinc finger protein 606 |
| chr4_-_119327822 | 0.07 |
ENSRNOT00000012645
|
Arhgap25
|
Rho GTPase activating protein 25 |
| chr12_+_2140203 | 0.07 |
ENSRNOT00000084906
|
Camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr11_+_34865532 | 0.07 |
ENSRNOT00000050342
|
Dyrk1a
|
dual specificity tyrosine phosphorylation regulated kinase 1A |
| chr9_-_63641400 | 0.06 |
ENSRNOT00000087684
|
Satb2
|
SATB homeobox 2 |
| chr2_+_104020955 | 0.06 |
ENSRNOT00000045586
|
Mtfr1
|
mitochondrial fission regulator 1 |
| chr2_-_198706428 | 0.06 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr14_+_39663421 | 0.06 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr2_-_149444548 | 0.06 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr4_+_29535852 | 0.06 |
ENSRNOT00000087619
|
NEWGENE_621351
|
collagen, type I, alpha 2 |
| chr7_-_120770435 | 0.06 |
ENSRNOT00000077000
|
Ddx17
|
DEAD-box helicase 17 |
| chr10_+_36715565 | 0.06 |
ENSRNOT00000005048
|
Clk4
|
CDC-like kinase 4 |
| chrX_+_11050121 | 0.06 |
ENSRNOT00000093665
|
Med14
|
mediator complex subunit 14 |
| chr6_-_51498337 | 0.06 |
ENSRNOT00000012487
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr4_+_180291389 | 0.06 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr1_+_164706276 | 0.06 |
ENSRNOT00000024311
|
Olr36
|
olfactory receptor 36 |
| chr17_-_89163113 | 0.06 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr4_+_66090209 | 0.06 |
ENSRNOT00000031193
|
Ttc26
|
tetratricopeptide repeat domain 26 |
| chr3_+_54253949 | 0.06 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr6_-_114476723 | 0.06 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr6_+_135680615 | 0.06 |
ENSRNOT00000077770
|
Traf3
|
Tnf receptor-associated factor 3 |
| chr1_-_275876329 | 0.06 |
ENSRNOT00000047903
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr18_+_59748444 | 0.06 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr14_-_28967980 | 0.06 |
ENSRNOT00000048175
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr10_+_54352270 | 0.05 |
ENSRNOT00000036752
|
Dhrs7c
|
dehydrogenase/reductase 7C |
| chr2_+_122877286 | 0.05 |
ENSRNOT00000033080
|
Arse
|
arylsulfatase E |
| chr1_-_120893997 | 0.05 |
ENSRNOT00000081028
|
AABR07003933.1
|
|
| chr20_-_4542073 | 0.05 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr18_+_30904498 | 0.05 |
ENSRNOT00000026969
|
Pcdhga11
|
protocadherin gamma subfamily A, 11 |
| chr17_+_89452814 | 0.05 |
ENSRNOT00000058760
|
RGD1561231
|
similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase like 1) |
| chr7_+_141666111 | 0.05 |
ENSRNOT00000083250
|
AABR07058886.1
|
|
| chr16_-_20562399 | 0.05 |
ENSRNOT00000031692
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr7_-_143967484 | 0.05 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr7_+_42304534 | 0.05 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr2_+_220432037 | 0.05 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr17_-_79085076 | 0.05 |
ENSRNOT00000057851
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr16_+_71629525 | 0.05 |
ENSRNOT00000035347
ENSRNOT00000088462 |
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr13_-_94859390 | 0.05 |
ENSRNOT00000005467
ENSRNOT00000079763 |
AABR07021840.1
|
|
| chr13_+_43818010 | 0.05 |
ENSRNOT00000005532
|
LOC100909664
|
centrosomal protein of 170 kDa-like |
| chr13_-_49828720 | 0.05 |
ENSRNOT00000012984
|
Mdm4
|
MDM4, p53 regulator |
| chr1_-_278042312 | 0.05 |
ENSRNOT00000018693
|
Ablim1
|
actin-binding LIM protein 1 |
| chr8_+_61671513 | 0.05 |
ENSRNOT00000091128
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr4_-_66955732 | 0.05 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr16_-_32421005 | 0.05 |
ENSRNOT00000082712
|
Nek1
|
NIMA-related kinase 1 |
| chrX_+_131381134 | 0.05 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr18_+_30869628 | 0.05 |
ENSRNOT00000060470
|
Pcdhgb4
|
protocadherin gamma subfamily B, 4 |
| chr20_+_4357733 | 0.05 |
ENSRNOT00000000509
|
Pbx2
|
PBX homeobox 2 |
| chr13_-_67206688 | 0.05 |
ENSRNOT00000003630
ENSRNOT00000090693 |
Pla2g4a
|
phospholipase A2 group IVA |
| chr3_+_100769839 | 0.05 |
ENSRNOT00000077703
|
Bdnf
|
brain-derived neurotrophic factor |
| chr19_-_22194740 | 0.05 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr1_+_260093641 | 0.05 |
ENSRNOT00000019521
|
Ccnj
|
cyclin J |
| chr13_-_102857551 | 0.05 |
ENSRNOT00000080309
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr1_-_236729412 | 0.05 |
ENSRNOT00000054794
|
Prune2
|
prune homolog 2 |
| chr17_+_63635086 | 0.04 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr18_+_24849545 | 0.04 |
ENSRNOT00000061011
ENSRNOT00000085516 |
Iws1
|
IWS1, SUPT6H interacting protein |
| chr16_-_23975061 | 0.04 |
ENSRNOT00000090726
|
Nat1
|
N-acetyltransferase 1 |
| chr9_-_20154077 | 0.04 |
ENSRNOT00000082904
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr6_+_123034304 | 0.04 |
ENSRNOT00000078938
|
Cpg1
|
candidate plasticity gene 1 |
| chr7_-_52404774 | 0.04 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chrX_-_135419704 | 0.04 |
ENSRNOT00000044435
|
Zfp280c
|
zinc finger protein 280C |
| chr13_+_6679368 | 0.04 |
ENSRNOT00000067198
ENSRNOT00000092965 |
Cntnap5c
|
contactin associated protein-like 5C |
| chr20_+_34258791 | 0.04 |
ENSRNOT00000000468
|
Slc35f1
|
solute carrier family 35, member F1 |
| chr5_-_119564846 | 0.04 |
ENSRNOT00000012977
|
Cyp2j4
|
cytochrome P450, family 2, subfamily j, polypeptide 4 |
| chr19_+_39229754 | 0.04 |
ENSRNOT00000050612
|
Vps4a
|
vacuolar protein sorting 4 homolog A |
| chr9_+_71915421 | 0.04 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr17_-_76281660 | 0.04 |
ENSRNOT00000058070
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr2_+_86891092 | 0.04 |
ENSRNOT00000077162
ENSRNOT00000083066 |
LOC100360380
|
zinc finger protein 457-like |
| chr11_-_11585078 | 0.04 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr15_-_52399074 | 0.04 |
ENSRNOT00000018440
|
Xpo7
|
exportin 7 |
| chr1_+_13595295 | 0.04 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr5_+_139790395 | 0.04 |
ENSRNOT00000015033
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr8_-_78397123 | 0.04 |
ENSRNOT00000087270
ENSRNOT00000084925 |
Tcf12
|
transcription factor 12 |
| chr11_+_15081774 | 0.04 |
ENSRNOT00000009911
|
LOC108348050
|
ubiquitin carboxyl-terminal hydrolase 25-like |
| chr5_-_166116516 | 0.04 |
ENSRNOT00000079919
ENSRNOT00000080888 |
Kif1b
|
kinesin family member 1B |
| chr7_-_114573900 | 0.04 |
ENSRNOT00000011219
|
Ptk2
|
protein tyrosine kinase 2 |
| chrX_+_84064427 | 0.04 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr9_+_60039297 | 0.04 |
ENSRNOT00000016262
|
Slc39a10
|
solute carrier family 39 member 10 |
| chr2_+_240642847 | 0.04 |
ENSRNOT00000079694
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chrX_-_14972675 | 0.04 |
ENSRNOT00000079664
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr17_+_54298409 | 0.04 |
ENSRNOT00000084919
|
Arhgap12
|
Rho GTPase activating protein 12 |
| chr8_+_106816152 | 0.04 |
ENSRNOT00000047613
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr4_+_31229913 | 0.04 |
ENSRNOT00000077134
ENSRNOT00000087897 |
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr3_+_129885826 | 0.03 |
ENSRNOT00000009743
|
Slx4ip
|
SLX4 interacting protein |
| chr12_-_10967326 | 0.03 |
ENSRNOT00000042095
|
LOC100912059
|
ubiquitin carboxyl-terminal hydrolase 12-like |
| chr17_-_87826421 | 0.03 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr16_+_70081035 | 0.03 |
ENSRNOT00000016098
|
LOC100912059
|
ubiquitin carboxyl-terminal hydrolase 12-like |
| chr2_+_248398917 | 0.03 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr5_-_60655985 | 0.03 |
ENSRNOT00000093103
|
Fbxo10
|
F-box protein 10 |
| chr3_+_48106099 | 0.03 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr20_+_31339787 | 0.03 |
ENSRNOT00000082463
|
Aifm2
|
apoptosis inducing factor, mitochondria associated 2 |
| chr7_+_12022285 | 0.03 |
ENSRNOT00000024080
|
Rexo1
|
RNA exonuclease 1 homolog |
| chr6_+_64789940 | 0.03 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr2_+_30685840 | 0.03 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chr3_-_138683318 | 0.03 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr14_+_69800156 | 0.03 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr6_+_104291071 | 0.03 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr2_-_147392062 | 0.03 |
ENSRNOT00000021535
|
Tm4sf1
|
transmembrane 4 L six family member 1 |
| chr7_-_29233392 | 0.03 |
ENSRNOT00000064241
|
Spic
|
Spi-C transcription factor |
| chr15_+_62406873 | 0.03 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr4_+_88694583 | 0.03 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr1_-_67134827 | 0.03 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr2_+_43271092 | 0.03 |
ENSRNOT00000017571
|
Mier3
|
mesoderm induction early response 1, family member 3 |
| chr4_+_3959640 | 0.03 |
ENSRNOT00000009439
|
Paxip1
|
PAX interacting protein 1 |
| chr10_-_62184874 | 0.03 |
ENSRNOT00000004229
|
Rpa1
|
replication protein A1 |
| chr1_-_53802658 | 0.03 |
ENSRNOT00000032667
|
Afdn
|
afadin, adherens junction formation factor |
| chr13_-_50916982 | 0.03 |
ENSRNOT00000004408
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr3_-_15278645 | 0.03 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr17_-_84247038 | 0.03 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr10_-_82117109 | 0.03 |
ENSRNOT00000079711
|
Abcc3
|
ATP binding cassette subfamily C member 3 |
| chr5_+_22392732 | 0.03 |
ENSRNOT00000087182
|
Clvs1
|
clavesin 1 |
| chr19_+_40855433 | 0.03 |
ENSRNOT00000023823
|
Il34
|
interleukin 34 |
| chr2_-_60657712 | 0.03 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr1_+_79981355 | 0.03 |
ENSRNOT00000020134
|
Dmwd
|
dystrophia myotonica, WD repeat containing |
| chr6_+_58468155 | 0.03 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr8_+_62283336 | 0.03 |
ENSRNOT00000025410
|
Rpp25
|
ribonuclease P/MRP 25 subunit |
| chr15_-_88670349 | 0.03 |
ENSRNOT00000012881
|
Rnf219
|
ring finger protein 219 |
| chr1_+_15782477 | 0.03 |
ENSRNOT00000088025
ENSRNOT00000089966 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr7_-_143353925 | 0.03 |
ENSRNOT00000068533
|
Krt71
|
keratin 71 |
| chr8_+_107875991 | 0.03 |
ENSRNOT00000090299
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr7_-_134482607 | 0.03 |
ENSRNOT00000081104
|
Gxylt1
|
glucoside xylosyltransferase 1 |
| chr1_+_88113445 | 0.02 |
ENSRNOT00000056955
|
Ggn
|
gametogenetin |
| chrX_+_9956370 | 0.02 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr14_+_7171613 | 0.02 |
ENSRNOT00000081600
|
Klhl8
|
kelch-like family member 8 |
| chr3_-_57607683 | 0.02 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr9_+_10941613 | 0.02 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr10_+_94260197 | 0.02 |
ENSRNOT00000063973
|
Taco1
|
translational activator of cytochrome c oxidase I |
| chr13_-_98078946 | 0.02 |
ENSRNOT00000035875
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr17_+_23661429 | 0.02 |
ENSRNOT00000046523
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr13_-_90405591 | 0.02 |
ENSRNOT00000006849
|
Vangl2
|
VANGL planar cell polarity protein 2 |
| chr9_+_66888393 | 0.02 |
ENSRNOT00000023536
|
Carf
|
calcium responsive transcription factor |
| chr2_+_219598162 | 0.02 |
ENSRNOT00000020297
|
Lrrc39
|
leucine rich repeat containing 39 |
| chr6_+_97168453 | 0.02 |
ENSRNOT00000085790
|
Syt16
|
synaptotagmin 16 |
| chr3_+_152626757 | 0.02 |
ENSRNOT00000086425
|
Aar2
|
AAR2 splicing factor homolog |
| chr14_+_83897117 | 0.02 |
ENSRNOT00000026569
|
Morc2
|
MORC family CW-type zinc finger 2 |
| chr8_+_107229832 | 0.02 |
ENSRNOT00000045821
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr9_-_78607307 | 0.02 |
ENSRNOT00000057672
|
Abca12
|
ATP binding cassette subfamily A member 12 |
| chr2_+_102685513 | 0.02 |
ENSRNOT00000033940
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr6_-_61405195 | 0.02 |
ENSRNOT00000008655
|
Lrrn3
|
leucine rich repeat neuronal 3 |
| chr17_-_56068125 | 0.02 |
ENSRNOT00000022462
|
Mtpap
|
mitochondrial poly(A) polymerase |
| chr11_+_65743892 | 0.02 |
ENSRNOT00000085226
|
AC106292.2
|
|
| chr10_+_90230441 | 0.02 |
ENSRNOT00000082722
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr2_+_243701962 | 0.02 |
ENSRNOT00000016891
|
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx5_Lmx1b_Lhx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.1 | GO:0032679 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:1900019 | activation of phospholipase D activity(GO:0031584) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.2 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.0 | GO:0036258 | multivesicular body assembly(GO:0036258) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0070668 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.3 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.0 | 0.1 | GO:0044307 | germinal vesicle(GO:0042585) dendritic branch(GO:0044307) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |