Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
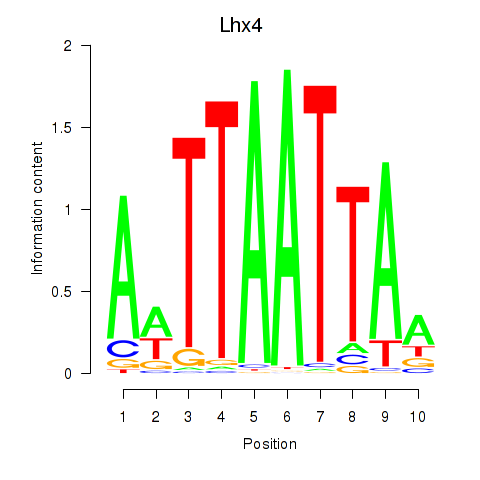
Results for Lhx4
Z-value: 0.23
Transcription factors associated with Lhx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx4
|
ENSRNOG00000003595 | LIM homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx4 | rn6_v1_chr13_-_73390393_73390543 | -0.36 | 5.5e-01 | Click! |
Activity profile of Lhx4 motif
Sorted Z-values of Lhx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_47677720 | 0.17 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr17_+_63635086 | 0.15 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr10_+_61685645 | 0.11 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr19_+_25043680 | 0.11 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr2_+_127525285 | 0.10 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr12_-_46493203 | 0.10 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr16_+_84465656 | 0.10 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr3_+_14889510 | 0.09 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr13_+_95589668 | 0.09 |
ENSRNOT00000005849
|
Zfp238
|
zinc finger protein 238 |
| chr12_+_25435567 | 0.08 |
ENSRNOT00000092982
|
Gtf2i
|
general transcription factor II I |
| chr1_-_166912524 | 0.08 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr15_-_54528480 | 0.07 |
ENSRNOT00000066888
|
Fndc3a
|
fibronectin type III domain containing 3a |
| chr4_-_18396035 | 0.06 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr11_-_62451149 | 0.06 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_-_49844547 | 0.06 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr17_+_9596957 | 0.06 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr2_+_198303168 | 0.06 |
ENSRNOT00000056262
|
Mtmr11
|
myotubularin related protein 11 |
| chr8_+_133210473 | 0.06 |
ENSRNOT00000082774
|
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr9_+_67234303 | 0.06 |
ENSRNOT00000050179
|
Abi2
|
abl-interactor 2 |
| chr10_-_62184874 | 0.06 |
ENSRNOT00000004229
|
Rpa1
|
replication protein A1 |
| chr2_-_33025271 | 0.06 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_+_64789940 | 0.06 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr17_-_89923423 | 0.05 |
ENSRNOT00000076964
|
Acbd5
|
acyl-CoA binding domain containing 5 |
| chr11_+_34865532 | 0.05 |
ENSRNOT00000050342
|
Dyrk1a
|
dual specificity tyrosine phosphorylation regulated kinase 1A |
| chr8_-_39551700 | 0.05 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr18_+_30895831 | 0.05 |
ENSRNOT00000026985
|
Pcdhga10
|
protocadherin gamma subfamily A, 10 |
| chr1_-_88162583 | 0.05 |
ENSRNOT00000087411
|
Catsperg
|
cation channel sperm associated auxiliary subunit gamma |
| chr10_+_45659143 | 0.05 |
ENSRNOT00000058327
|
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr15_+_57505449 | 0.05 |
ENSRNOT00000064945
|
Siah3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr4_+_181315444 | 0.05 |
ENSRNOT00000044147
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr10_-_16731898 | 0.05 |
ENSRNOT00000028186
|
Crebrf
|
CREB3 regulatory factor |
| chr11_-_4397361 | 0.05 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr2_-_32518643 | 0.05 |
ENSRNOT00000061032
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr15_-_93765498 | 0.05 |
ENSRNOT00000093297
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr2_-_35870578 | 0.05 |
ENSRNOT00000066069
|
Rnf180
|
ring finger protein 180 |
| chr4_+_66090209 | 0.05 |
ENSRNOT00000031193
|
Ttc26
|
tetratricopeptide repeat domain 26 |
| chrX_+_71540895 | 0.04 |
ENSRNOT00000004692
ENSRNOT00000082967 |
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr18_-_43945273 | 0.04 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr8_-_78397123 | 0.04 |
ENSRNOT00000087270
ENSRNOT00000084925 |
Tcf12
|
transcription factor 12 |
| chr18_+_16146447 | 0.04 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr13_-_83425641 | 0.04 |
ENSRNOT00000063870
|
Tbx19
|
T-box 19 |
| chr5_+_155934490 | 0.04 |
ENSRNOT00000077933
|
LOC100911993
|
ubiquitin carboxyl-terminal hydrolase 48-like |
| chr7_-_114590119 | 0.04 |
ENSRNOT00000079599
|
Ptk2
|
protein tyrosine kinase 2 |
| chr2_+_136993208 | 0.04 |
ENSRNOT00000040187
ENSRNOT00000066542 |
Pcdh10
|
protocadherin 10 |
| chr1_-_128287151 | 0.04 |
ENSRNOT00000084946
ENSRNOT00000089723 |
Mef2a
|
myocyte enhancer factor 2a |
| chr7_+_42304534 | 0.04 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr14_-_77810147 | 0.04 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chr13_-_83457888 | 0.04 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr11_+_15081774 | 0.04 |
ENSRNOT00000009911
|
LOC108348050
|
ubiquitin carboxyl-terminal hydrolase 25-like |
| chr4_-_55011415 | 0.04 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr2_-_149444548 | 0.04 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr8_-_7426611 | 0.04 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr15_+_62406873 | 0.04 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr11_-_11585078 | 0.04 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr2_+_165486910 | 0.04 |
ENSRNOT00000012982
|
LOC100362176
|
hypothetical protein LOC100362176 |
| chr17_-_78812111 | 0.04 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr18_+_24849545 | 0.03 |
ENSRNOT00000061011
ENSRNOT00000085516 |
Iws1
|
IWS1, SUPT6H interacting protein |
| chr8_+_82038967 | 0.03 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr2_+_4989295 | 0.03 |
ENSRNOT00000041541
|
Fam172a
|
family with sequence similarity 172, member A |
| chr8_-_39460844 | 0.03 |
ENSRNOT00000048875
|
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr5_-_16799776 | 0.03 |
ENSRNOT00000011721
|
Plag1
|
PLAG1 zinc finger |
| chr1_-_90520344 | 0.03 |
ENSRNOT00000078598
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr2_+_86996497 | 0.03 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr2_-_38110567 | 0.03 |
ENSRNOT00000072212
|
Ipo11
|
importin 11 |
| chr20_-_13994794 | 0.03 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr9_-_100592271 | 0.03 |
ENSRNOT00000086433
|
Hdlbp
|
high density lipoprotein binding protein |
| chr5_+_25725683 | 0.03 |
ENSRNOT00000087602
|
LOC679087
|
similar to swan |
| chr17_+_54298409 | 0.03 |
ENSRNOT00000084919
|
Arhgap12
|
Rho GTPase activating protein 12 |
| chr1_+_101603222 | 0.03 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr7_-_52404774 | 0.03 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr8_-_119012671 | 0.03 |
ENSRNOT00000028435
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr2_-_248789508 | 0.03 |
ENSRNOT00000090705
|
Pkn2
|
protein kinase N2 |
| chr17_-_15467320 | 0.03 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chr14_+_113867209 | 0.03 |
ENSRNOT00000067591
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr9_+_94324793 | 0.03 |
ENSRNOT00000092493
|
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr8_+_70317586 | 0.03 |
ENSRNOT00000016260
|
Dennd4a
|
DENN domain containing 4A |
| chr10_+_103206014 | 0.03 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chrX_-_138118696 | 0.03 |
ENSRNOT00000084130
|
Frmd7
|
FERM domain containing 7 |
| chr4_+_169161585 | 0.03 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr2_+_220432037 | 0.03 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr3_-_90751055 | 0.03 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr19_-_43192208 | 0.03 |
ENSRNOT00000025179
|
Exosc6
|
exosome component 6 |
| chr11_+_47146308 | 0.03 |
ENSRNOT00000002191
|
Cep97
|
centrosomal protein 97 |
| chr4_-_180234804 | 0.03 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr14_+_45982244 | 0.03 |
ENSRNOT00000076881
ENSRNOT00000002984 |
Rell1
|
RELT-like 1 |
| chr1_-_66237501 | 0.02 |
ENSRNOT00000073006
|
Zfp606
|
zinc finger protein 606 |
| chr9_+_73418607 | 0.02 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr1_-_53802658 | 0.02 |
ENSRNOT00000032667
|
Afdn
|
afadin, adherens junction formation factor |
| chr6_+_134844676 | 0.02 |
ENSRNOT00000007595
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr18_+_30840868 | 0.02 |
ENSRNOT00000027026
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr15_+_110114148 | 0.02 |
ENSRNOT00000006264
|
Itgbl1
|
integrin subunit beta like 1 |
| chr5_+_25253010 | 0.02 |
ENSRNOT00000061333
|
Rbm12b
|
RNA binding motif protein 12B |
| chr16_+_70081035 | 0.02 |
ENSRNOT00000016098
|
LOC100912059
|
ubiquitin carboxyl-terminal hydrolase 12-like |
| chr13_+_57243877 | 0.02 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr17_-_69862110 | 0.02 |
ENSRNOT00000058312
|
Akr1cl
|
aldo-keto reductase family 1, member C-like |
| chr8_+_103938520 | 0.02 |
ENSRNOT00000067835
|
Gk5
|
glycerol kinase 5 (putative) |
| chr7_+_122979021 | 0.02 |
ENSRNOT00000085707
|
Zc3h7b
|
zinc finger CCCH-type containing 7B |
| chr15_+_3996048 | 0.02 |
ENSRNOT00000078653
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr9_-_42839837 | 0.02 |
ENSRNOT00000038610
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr12_+_48677905 | 0.02 |
ENSRNOT00000083196
|
1700069L16Rik
|
RIKEN cDNA 1700069L16 gene |
| chr12_-_10967326 | 0.02 |
ENSRNOT00000042095
|
LOC100912059
|
ubiquitin carboxyl-terminal hydrolase 12-like |
| chr18_+_59748444 | 0.02 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr2_+_86996798 | 0.02 |
ENSRNOT00000070821
|
Zfp455
|
zinc finger protein 455 |
| chr8_-_21481735 | 0.02 |
ENSRNOT00000038069
|
Zfp426
|
zinc finger protein 426 |
| chr7_-_105592804 | 0.02 |
ENSRNOT00000006789
|
Adcy8
|
adenylate cyclase 8 |
| chr9_-_69878706 | 0.02 |
ENSRNOT00000035879
|
Ino80d
|
INO80 complex subunit D |
| chr7_+_120923274 | 0.02 |
ENSRNOT00000049247
|
Gtpbp1
|
GTP binding protein 1 |
| chr13_-_76049363 | 0.02 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr6_-_108660063 | 0.02 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr2_+_102685513 | 0.02 |
ENSRNOT00000033940
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr3_-_67787990 | 0.02 |
ENSRNOT00000064851
|
Nckap1
|
NCK-associated protein 1 |
| chr2_-_186245342 | 0.02 |
ENSRNOT00000057062
ENSRNOT00000022292 |
Dclk2
|
doublecortin-like kinase 2 |
| chrX_-_111191932 | 0.02 |
ENSRNOT00000088050
ENSRNOT00000083613 |
Morc4
|
MORC family CW-type zinc finger 4 |
| chr8_+_26432514 | 0.02 |
ENSRNOT00000081574
|
Sept7
|
septin 7 |
| chr16_+_6048004 | 0.02 |
ENSRNOT00000020750
|
Actr8
|
ARP8 actin-related protein 8 homolog |
| chr3_-_15278645 | 0.02 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr1_+_101599018 | 0.02 |
ENSRNOT00000028494
|
Fut1
|
fucosyltransferase 1 |
| chr14_+_39964588 | 0.02 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr3_+_8430829 | 0.02 |
ENSRNOT00000090440
ENSRNOT00000060969 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr2_-_186245163 | 0.02 |
ENSRNOT00000089339
|
Dclk2
|
doublecortin-like kinase 2 |
| chr20_+_3155652 | 0.02 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr7_-_140291620 | 0.02 |
ENSRNOT00000088323
|
Adcy6
|
adenylate cyclase 6 |
| chr16_+_71629525 | 0.02 |
ENSRNOT00000035347
ENSRNOT00000088462 |
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr20_-_4542073 | 0.01 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr1_+_198214797 | 0.01 |
ENSRNOT00000068543
|
Tbx6
|
T-box 6 |
| chr5_-_136166786 | 0.01 |
ENSRNOT00000026201
|
Rnf220
|
ring finger protein 220 |
| chr18_+_40853988 | 0.01 |
ENSRNOT00000091759
|
AABR07031963.1
|
uncharacterized protein LOC317165 |
| chr17_-_76281660 | 0.01 |
ENSRNOT00000058070
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr9_+_79630604 | 0.01 |
ENSRNOT00000021539
ENSRNOT00000083979 |
Tmem169
|
transmembrane protein 169 |
| chr6_+_18880737 | 0.01 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr5_+_156587093 | 0.01 |
ENSRNOT00000019696
|
Hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr11_+_47061354 | 0.01 |
ENSRNOT00000039997
|
Pcnp
|
PEST proteolytic signal containing nuclear protein |
| chr1_+_260093641 | 0.01 |
ENSRNOT00000019521
|
Ccnj
|
cyclin J |
| chr4_-_72143748 | 0.01 |
ENSRNOT00000024428
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr9_-_16643182 | 0.01 |
ENSRNOT00000024266
|
LOC108348250
|
cullin-7 |
| chr3_+_100769839 | 0.01 |
ENSRNOT00000077703
|
Bdnf
|
brain-derived neurotrophic factor |
| chr6_+_48452369 | 0.01 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chrX_+_84064427 | 0.01 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr3_+_56766475 | 0.01 |
ENSRNOT00000078819
|
Sp5
|
Sp5 transcription factor |
| chr10_+_67810810 | 0.01 |
ENSRNOT00000079156
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr6_+_44225233 | 0.01 |
ENSRNOT00000066593
|
Kidins220
|
kinase D-interacting substrate 220 |
| chr3_-_57607683 | 0.01 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr5_+_138605793 | 0.01 |
ENSRNOT00000083754
|
Foxj3
|
forkhead box J3 |
| chr12_-_23727535 | 0.01 |
ENSRNOT00000085911
ENSRNOT00000001950 |
Dtx2
|
deltex E3 ubiquitin ligase 2 |
| chr8_+_61671513 | 0.01 |
ENSRNOT00000091128
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr8_+_106586689 | 0.01 |
ENSRNOT00000080434
|
Copb2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr4_-_148437961 | 0.01 |
ENSRNOT00000082907
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr14_+_113530470 | 0.01 |
ENSRNOT00000004919
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr1_-_56683731 | 0.01 |
ENSRNOT00000014552
|
Thbs2
|
thrombospondin 2 |
| chr2_+_118831350 | 0.01 |
ENSRNOT00000083720
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr4_+_68635581 | 0.01 |
ENSRNOT00000016217
|
Ssbp1
|
single stranded DNA binding protein 1 |
| chr1_+_72380711 | 0.01 |
ENSRNOT00000022236
|
Fiz1
|
FLT3-interacting zinc finger 1 |
| chr1_-_101819478 | 0.01 |
ENSRNOT00000056181
|
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr19_-_29968424 | 0.01 |
ENSRNOT00000024981
|
Inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr2_-_57935334 | 0.01 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr2_+_127489771 | 0.01 |
ENSRNOT00000093581
|
Intu
|
inturned planar cell polarity protein |
| chr6_-_67084234 | 0.01 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr1_-_38538987 | 0.01 |
ENSRNOT00000090406
ENSRNOT00000071275 |
LOC102551340
|
zinc finger protein 728-like |
| chr5_+_58995249 | 0.01 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr2_+_189857587 | 0.01 |
ENSRNOT00000048214
|
Ilf2
|
interleukin enhancer binding factor 2 |
| chr2_-_127781003 | 0.01 |
ENSRNOT00000050764
|
Mfsd8
|
major facilitator superfamily domain containing 8 |
| chr6_-_23291568 | 0.00 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr3_-_45169118 | 0.00 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr20_+_25990304 | 0.00 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr5_-_100970043 | 0.00 |
ENSRNOT00000013950
|
Zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr20_+_4357733 | 0.00 |
ENSRNOT00000000509
|
Pbx2
|
PBX homeobox 2 |
| chr8_+_117297670 | 0.00 |
ENSRNOT00000082628
|
Qars
|
glutaminyl-tRNA synthetase |
| chr2_-_157759819 | 0.00 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr2_-_227411964 | 0.00 |
ENSRNOT00000019931
|
Synpo2
|
synaptopodin 2 |
| chr1_+_164706276 | 0.00 |
ENSRNOT00000024311
|
Olr36
|
olfactory receptor 36 |
| chr2_-_198706428 | 0.00 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr9_-_14550625 | 0.00 |
ENSRNOT00000078227
|
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr10_+_67810655 | 0.00 |
ENSRNOT00000064285
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr14_-_44078897 | 0.00 |
ENSRNOT00000031792
|
N4bp2
|
NEDD4 binding protein 2 |
| chr2_+_226563050 | 0.00 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr4_-_184096806 | 0.00 |
ENSRNOT00000055433
|
LOC100362344
|
mKIAA1238 protein-like |
| chrX_+_144994139 | 0.00 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr6_-_111417813 | 0.00 |
ENSRNOT00000016324
ENSRNOT00000085458 |
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr14_-_81053905 | 0.00 |
ENSRNOT00000045068
ENSRNOT00000040215 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr1_+_79631668 | 0.00 |
ENSRNOT00000083546
ENSRNOT00000035286 |
Mill1
|
MHC I like leukocyte 1 |
| chr10_+_1834518 | 0.00 |
ENSRNOT00000061709
|
Gm1758
|
predicted gene 1758 |
| chr8_-_132790778 | 0.00 |
ENSRNOT00000008255
ENSRNOT00000091431 |
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr6_+_69971227 | 0.00 |
ENSRNOT00000075349
|
Foxg1
|
forkhead box G1 |
| chr15_+_37790141 | 0.00 |
ENSRNOT00000076392
ENSRNOT00000091953 |
Il17d
|
interleukin 17D |
| chr20_+_27366213 | 0.00 |
ENSRNOT00000000302
ENSRNOT00000057939 |
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr15_-_93748742 | 0.00 |
ENSRNOT00000093370
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr18_-_26656879 | 0.00 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr2_+_30685840 | 0.00 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chrX_+_4285857 | 0.00 |
ENSRNOT00000082159
|
AABR07036855.1
|
|
| chr3_+_17889972 | 0.00 |
ENSRNOT00000073021
|
AABR07051611.1
|
|
| chr6_+_2216623 | 0.00 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr12_-_24102084 | 0.00 |
ENSRNOT00000001962
|
Rhbdd2
|
rhomboid domain containing 2 |
| chr14_+_37116492 | 0.00 |
ENSRNOT00000002921
|
Sgcb
|
sarcoglycan, beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:2000458 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.0 | 0.1 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.0 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.0 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.0 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.0 | GO:0007172 | signal complex assembly(GO:0007172) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.0 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.0 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |