Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
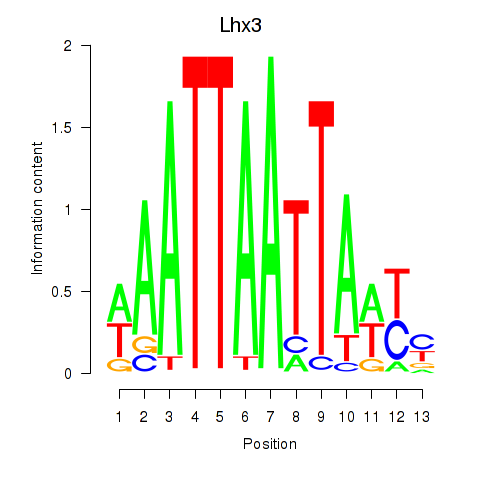
Results for Lhx3
Z-value: 0.34
Transcription factors associated with Lhx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx3
|
ENSRNOG00000018427 | LIM homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx3 | rn6_v1_chr3_-_3661810_3661810 | 0.95 | 1.2e-02 | Click! |
Activity profile of Lhx3 motif
Sorted Z-values of Lhx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_2174131 | 0.30 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr1_-_264975132 | 0.12 |
ENSRNOT00000021748
|
Lbx1
|
ladybird homeobox 1 |
| chr2_+_145174876 | 0.10 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr15_-_93765498 | 0.09 |
ENSRNOT00000093297
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chrX_-_40086870 | 0.08 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr9_+_73418607 | 0.06 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr8_+_19888667 | 0.06 |
ENSRNOT00000078593
|
Zfp317
|
zinc finger protein 317 |
| chr7_+_42304534 | 0.06 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr10_+_103206014 | 0.05 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr8_-_7426611 | 0.05 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr13_-_83457888 | 0.05 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr12_+_25435567 | 0.05 |
ENSRNOT00000092982
|
Gtf2i
|
general transcription factor II I |
| chr5_-_16799776 | 0.05 |
ENSRNOT00000011721
|
Plag1
|
PLAG1 zinc finger |
| chr1_+_238222521 | 0.05 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr19_-_37528011 | 0.05 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr9_+_66888393 | 0.05 |
ENSRNOT00000023536
|
Carf
|
calcium responsive transcription factor |
| chr8_-_119012671 | 0.04 |
ENSRNOT00000028435
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr17_-_89923423 | 0.04 |
ENSRNOT00000076964
|
Acbd5
|
acyl-CoA binding domain containing 5 |
| chr14_+_7026769 | 0.04 |
ENSRNOT00000071955
|
LOC100359907
|
secretory calcium-binding phosphoprotein proline-glutamine rich 1-like |
| chr18_-_43945273 | 0.04 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr18_+_16146447 | 0.04 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr1_+_101603222 | 0.04 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr2_-_149444548 | 0.04 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chrX_+_151103576 | 0.04 |
ENSRNOT00000015401
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr15_-_54528480 | 0.04 |
ENSRNOT00000066888
|
Fndc3a
|
fibronectin type III domain containing 3a |
| chr3_-_71845232 | 0.04 |
ENSRNOT00000078645
|
Calcrl
|
calcitonin receptor like receptor |
| chr2_+_86996497 | 0.03 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr2_+_95320283 | 0.03 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr9_-_88086488 | 0.03 |
ENSRNOT00000019579
|
Irs1
|
insulin receptor substrate 1 |
| chr13_+_84474319 | 0.03 |
ENSRNOT00000031367
ENSRNOT00000072244 ENSRNOT00000072897 ENSRNOT00000064168 ENSRNOT00000074954 ENSRNOT00000073696 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr14_+_36047144 | 0.03 |
ENSRNOT00000003088
|
Lnx1
|
ligand of numb-protein X 1 |
| chrX_-_29648359 | 0.03 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr6_+_64789940 | 0.02 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr6_+_2216623 | 0.02 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr20_+_3155652 | 0.02 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr1_-_101095594 | 0.02 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr1_-_78180216 | 0.02 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr10_+_61685645 | 0.02 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chrX_-_111191932 | 0.02 |
ENSRNOT00000088050
ENSRNOT00000083613 |
Morc4
|
MORC family CW-type zinc finger 4 |
| chr3_+_148108539 | 0.02 |
ENSRNOT00000010149
|
Rem1
|
RRAD and GEM like GTPase 1 |
| chr9_-_42839837 | 0.02 |
ENSRNOT00000038610
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr16_+_6048004 | 0.02 |
ENSRNOT00000020750
|
Actr8
|
ARP8 actin-related protein 8 homolog |
| chr15_-_57651041 | 0.02 |
ENSRNOT00000072138
|
Spert
|
spermatid associated |
| chr12_-_39641285 | 0.02 |
ENSRNOT00000001729
|
Anapc7
|
anaphase promoting complex subunit 7 |
| chr15_+_62406873 | 0.01 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr2_-_31753528 | 0.01 |
ENSRNOT00000075057
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr20_-_4542073 | 0.01 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr7_+_120923274 | 0.01 |
ENSRNOT00000049247
|
Gtpbp1
|
GTP binding protein 1 |
| chr4_+_181315444 | 0.01 |
ENSRNOT00000044147
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr2_+_86996798 | 0.01 |
ENSRNOT00000070821
|
Zfp455
|
zinc finger protein 455 |
| chr10_+_67810810 | 0.01 |
ENSRNOT00000079156
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr5_+_6373583 | 0.01 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr17_-_78812111 | 0.01 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr19_-_29968424 | 0.01 |
ENSRNOT00000024981
|
Inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr1_-_101819478 | 0.01 |
ENSRNOT00000056181
|
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr4_-_148437961 | 0.01 |
ENSRNOT00000082907
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr2_+_30685840 | 0.01 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chr9_+_73378057 | 0.01 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr7_-_105592804 | 0.01 |
ENSRNOT00000006789
|
Adcy8
|
adenylate cyclase 8 |
| chr11_+_47061354 | 0.01 |
ENSRNOT00000039997
|
Pcnp
|
PEST proteolytic signal containing nuclear protein |
| chr8_+_117297670 | 0.01 |
ENSRNOT00000082628
|
Qars
|
glutaminyl-tRNA synthetase |
| chr14_+_39964588 | 0.01 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr10_+_67810655 | 0.01 |
ENSRNOT00000064285
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr18_+_59748444 | 0.01 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr16_+_23447366 | 0.01 |
ENSRNOT00000068629
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr11_-_60547201 | 0.01 |
ENSRNOT00000093151
|
Btla
|
B and T lymphocyte associated |
| chr9_-_121725716 | 0.01 |
ENSRNOT00000087405
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr15_-_93307420 | 0.01 |
ENSRNOT00000012195
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr3_+_102925471 | 0.00 |
ENSRNOT00000040364
|
Olr772
|
olfactory receptor 772 |
| chr18_+_55666027 | 0.00 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chr5_-_136166786 | 0.00 |
ENSRNOT00000026201
|
Rnf220
|
ring finger protein 220 |
| chr6_+_18880737 | 0.00 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr7_-_140291620 | 0.00 |
ENSRNOT00000088323
|
Adcy6
|
adenylate cyclase 6 |
| chr3_+_119561290 | 0.00 |
ENSRNOT00000015843
|
Blvra
|
biliverdin reductase A |
| chr1_+_79631668 | 0.00 |
ENSRNOT00000083546
ENSRNOT00000035286 |
Mill1
|
MHC I like leukocyte 1 |
| chr9_+_38536920 | 0.00 |
ENSRNOT00000060452
|
Prim2
|
primase (DNA) subunit 2 |
| chrX_-_9999401 | 0.00 |
ENSRNOT00000060992
|
Gpr82
|
G protein-coupled receptor 82 |
| chr17_+_53965562 | 0.00 |
ENSRNOT00000090387
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr2_-_233743866 | 0.00 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0070662 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.0 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.0 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |