Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
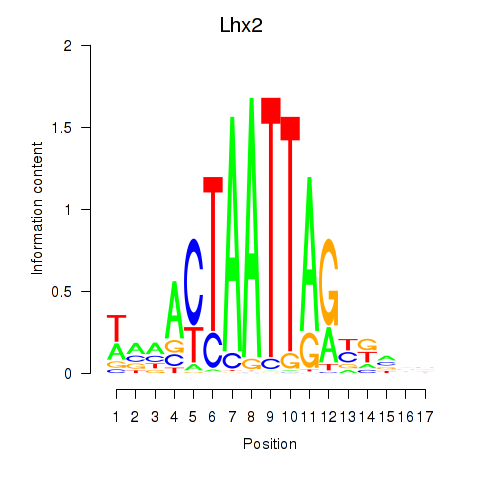
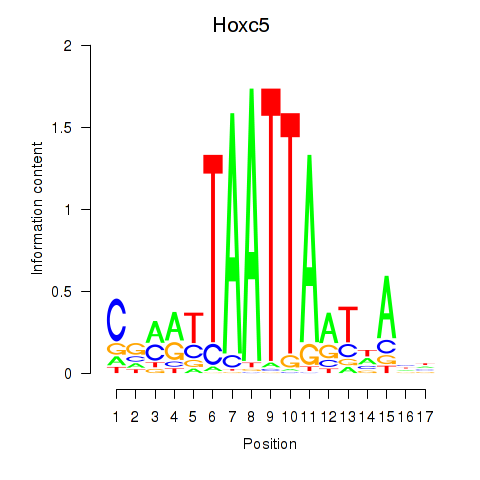
Results for Lhx2_Hoxc5
Z-value: 0.88
Transcription factors associated with Lhx2_Hoxc5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx2
|
ENSRNOG00000010551 | LIM homeobox 2 |
|
Hoxc5
|
ENSRNOG00000016598 | homeo box C5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc5 | rn6_v1_chr7_+_144628120_144628120 | -0.26 | 6.8e-01 | Click! |
| Lhx2 | rn6_v1_chr3_+_22640545_22640604 | -0.25 | 6.8e-01 | Click! |
Activity profile of Lhx2_Hoxc5 motif
Sorted Z-values of Lhx2_Hoxc5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrM_+_9870 | 0.57 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr5_+_6373583 | 0.54 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr8_-_104155775 | 0.48 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chrM_+_11736 | 0.47 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr13_-_83457888 | 0.47 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr15_-_93765498 | 0.44 |
ENSRNOT00000093297
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr1_+_15834779 | 0.39 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr15_+_87722221 | 0.36 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr1_+_61786900 | 0.35 |
ENSRNOT00000090287
|
AABR07001905.1
|
|
| chr13_+_91054974 | 0.35 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr2_-_149444548 | 0.33 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr15_-_42794279 | 0.32 |
ENSRNOT00000023385
|
Ephx2
|
epoxide hydrolase 2 |
| chr7_+_20262680 | 0.32 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr1_+_252409268 | 0.32 |
ENSRNOT00000026219
|
Lipm
|
lipase, family member M |
| chr6_+_86785771 | 0.32 |
ENSRNOT00000066702
|
Prpf39
|
pre-mRNA processing factor 39 |
| chr7_-_116106368 | 0.31 |
ENSRNOT00000035678
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr1_+_140998240 | 0.31 |
ENSRNOT00000023506
ENSRNOT00000090897 |
Abhd2
|
abhydrolase domain containing 2 |
| chr2_+_23289374 | 0.30 |
ENSRNOT00000090666
ENSRNOT00000032783 |
Dmgdh
|
dimethylglycine dehydrogenase |
| chr1_-_78180216 | 0.30 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr10_-_34242985 | 0.30 |
ENSRNOT00000046438
|
RGD1559575
|
similar to novel protein |
| chr10_-_16752205 | 0.25 |
ENSRNOT00000079462
|
Crebrf
|
CREB3 regulatory factor |
| chrX_-_70460536 | 0.25 |
ENSRNOT00000076824
|
Pdzd11
|
PDZ domain containing 11 |
| chr13_+_96303703 | 0.25 |
ENSRNOT00000084718
ENSRNOT00000029723 |
Efcab2
|
EF-hand calcium binding domain 2 |
| chr15_+_4240203 | 0.25 |
ENSRNOT00000010178
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr8_+_2604962 | 0.24 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chrX_+_84064427 | 0.24 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chrM_+_3904 | 0.23 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chrX_+_14019961 | 0.23 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr1_+_72956026 | 0.22 |
ENSRNOT00000031462
|
Rdh13
|
retinol dehydrogenase 13 |
| chr6_-_108660063 | 0.22 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr17_-_52477575 | 0.22 |
ENSRNOT00000081290
|
Gli3
|
GLI family zinc finger 3 |
| chr15_+_36809361 | 0.22 |
ENSRNOT00000076667
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr2_+_127489771 | 0.22 |
ENSRNOT00000093581
|
Intu
|
inturned planar cell polarity protein |
| chr17_-_84247038 | 0.22 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr7_-_80457816 | 0.21 |
ENSRNOT00000039430
|
AABR07057617.1
|
|
| chr4_-_16654811 | 0.21 |
ENSRNOT00000008637
|
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr4_+_61814974 | 0.21 |
ENSRNOT00000074951
|
Akr1b10
|
aldo-keto reductase family 1 member B10 |
| chr11_+_38727048 | 0.20 |
ENSRNOT00000081537
|
LOC103690343
|
zinc finger protein 260-like |
| chr3_-_90751055 | 0.20 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr2_+_187447501 | 0.20 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr6_+_76349362 | 0.20 |
ENSRNOT00000043224
|
Aldoart2
|
aldolase 1 A retrogene 2 |
| chr4_+_2053712 | 0.19 |
ENSRNOT00000045086
|
Rnf32
|
ring finger protein 32 |
| chr4_-_165456677 | 0.19 |
ENSRNOT00000082207
|
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chr7_-_76035096 | 0.19 |
ENSRNOT00000072255
|
AABR07057530.1
|
|
| chr6_-_91581262 | 0.19 |
ENSRNOT00000034507
|
AABR07064716.1
|
|
| chr1_-_166919302 | 0.19 |
ENSRNOT00000026925
|
Folr2
|
folate receptor beta |
| chr18_+_30487264 | 0.18 |
ENSRNOT00000040125
|
Pcdhb10
|
protocadherin beta 10 |
| chr2_-_157035483 | 0.18 |
ENSRNOT00000076491
|
LOC304239
|
similar to RalA binding protein 1 |
| chr17_+_45175121 | 0.18 |
ENSRNOT00000080417
|
Nkapl
|
NFKB activating protein-like |
| chr10_+_29289203 | 0.18 |
ENSRNOT00000067013
|
Pwwp2a
|
PWWP domain containing 2A |
| chr8_-_21492801 | 0.18 |
ENSRNOT00000077465
|
Zfp426
|
zinc finger protein 426 |
| chr1_-_261179790 | 0.18 |
ENSRNOT00000074420
ENSRNOT00000072073 |
Exosc1
|
exosome component 1 |
| chr1_-_38538987 | 0.18 |
ENSRNOT00000090406
ENSRNOT00000071275 |
LOC102551340
|
zinc finger protein 728-like |
| chr3_-_25212049 | 0.17 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr1_+_66959610 | 0.17 |
ENSRNOT00000072122
ENSRNOT00000045994 |
Vom1r48
|
vomeronasal 1 receptor 48 |
| chr2_-_34452895 | 0.17 |
ENSRNOT00000079385
|
AABR07007905.2
|
|
| chr2_+_118547190 | 0.17 |
ENSRNOT00000083676
ENSRNOT00000090301 ENSRNOT00000013410 |
Kcnmb2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr2_+_66940057 | 0.16 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr1_+_169145445 | 0.16 |
ENSRNOT00000034019
|
LOC499219
|
hypothetical protein LOC499219 |
| chrM_+_9451 | 0.16 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr8_+_12355767 | 0.16 |
ENSRNOT00000068445
|
Fam76b
|
family with sequence similarity 76, member B |
| chr7_+_15785410 | 0.16 |
ENSRNOT00000082664
ENSRNOT00000073235 |
Zfp955a
|
zinc finger protein 955A |
| chr13_-_67206688 | 0.16 |
ENSRNOT00000003630
ENSRNOT00000090693 |
Pla2g4a
|
phospholipase A2 group IVA |
| chr1_+_280103695 | 0.15 |
ENSRNOT00000065289
|
Eno4
|
enolase family member 4 |
| chrX_+_908044 | 0.15 |
ENSRNOT00000072087
|
Zfp300
|
zinc finger protein 300 |
| chr20_-_13994794 | 0.15 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr4_-_22424862 | 0.15 |
ENSRNOT00000082359
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr5_-_12172009 | 0.15 |
ENSRNOT00000061903
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr4_+_27509221 | 0.15 |
ENSRNOT00000067954
|
Ankib1
|
ankyrin repeat and IBR domain containing 1 |
| chr4_+_88694583 | 0.15 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr7_+_141666111 | 0.15 |
ENSRNOT00000083250
|
AABR07058886.1
|
|
| chr5_+_24410863 | 0.15 |
ENSRNOT00000010591
|
Tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr16_-_81714346 | 0.15 |
ENSRNOT00000092552
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr15_+_33885106 | 0.14 |
ENSRNOT00000024483
|
Dhrs2
|
dehydrogenase/reductase 2 |
| chr8_-_96547568 | 0.14 |
ENSRNOT00000078343
|
RGD1560775
|
similar to RIKEN cDNA 4930579C12 gene |
| chr7_+_91384187 | 0.14 |
ENSRNOT00000005828
|
Utp23
|
UTP23, small subunit processome component |
| chr5_-_168734296 | 0.14 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr9_+_30419001 | 0.14 |
ENSRNOT00000018116
|
Col9a1
|
collagen type IX alpha 1 chain |
| chrM_+_8599 | 0.14 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr2_+_86891092 | 0.14 |
ENSRNOT00000077162
ENSRNOT00000083066 |
LOC100360380
|
zinc finger protein 457-like |
| chr18_-_6587080 | 0.14 |
ENSRNOT00000040815
|
LOC103694404
|
60S ribosomal protein L39 |
| chr2_+_86996497 | 0.14 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr4_+_31229913 | 0.14 |
ENSRNOT00000077134
ENSRNOT00000087897 |
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr16_+_22979444 | 0.14 |
ENSRNOT00000017822
|
Csgalnact1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr10_+_36715565 | 0.14 |
ENSRNOT00000005048
|
Clk4
|
CDC-like kinase 4 |
| chr1_-_66237501 | 0.14 |
ENSRNOT00000073006
|
Zfp606
|
zinc finger protein 606 |
| chr6_+_86823684 | 0.14 |
ENSRNOT00000086081
ENSRNOT00000006574 |
Fancm
|
Fanconi anemia, complementation group M |
| chr4_+_147832136 | 0.13 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr19_-_21970103 | 0.13 |
ENSRNOT00000074210
|
AABR07043115.1
|
|
| chr10_+_56445647 | 0.13 |
ENSRNOT00000056870
|
Tmem256
|
transmembrane protein 256 |
| chr1_-_267463694 | 0.13 |
ENSRNOT00000084851
ENSRNOT00000016645 |
Col17a1
|
collagen type XVII alpha 1 chain |
| chr1_+_253221812 | 0.13 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chr16_-_7412150 | 0.13 |
ENSRNOT00000047252
|
RGD1564325
|
similar to ribosomal protein S24 |
| chr8_+_96551245 | 0.13 |
ENSRNOT00000039850
|
Bcl2a1
|
BCL2-related protein A1 |
| chr1_-_214202853 | 0.13 |
ENSRNOT00000022954
|
Lmntd2
|
lamin tail domain containing 2 |
| chr3_-_71798531 | 0.12 |
ENSRNOT00000088170
|
Calcrl
|
calcitonin receptor like receptor |
| chr14_+_38192446 | 0.12 |
ENSRNOT00000090242
|
Nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr9_-_95143092 | 0.12 |
ENSRNOT00000064171
ENSRNOT00000085325 |
Usp40
|
ubiquitin specific peptidase 40 |
| chr11_+_88095170 | 0.12 |
ENSRNOT00000041557
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr12_-_20276121 | 0.12 |
ENSRNOT00000065873
|
LOC685157
|
similar to paired immunoglobin-like type 2 receptor beta |
| chr12_-_24775891 | 0.12 |
ENSRNOT00000074851
|
Wbscr27
|
Williams Beuren syndrome chromosome region 27 |
| chr7_+_78558701 | 0.12 |
ENSRNOT00000006393
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chrX_-_158288109 | 0.12 |
ENSRNOT00000072527
|
LOC103690175
|
PHD finger protein 6 |
| chr19_-_37796089 | 0.12 |
ENSRNOT00000024891
|
Ranbp10
|
RAN binding protein 10 |
| chrX_+_156873849 | 0.12 |
ENSRNOT00000085410
|
Arhgap4
|
Rho GTPase activating protein 4 |
| chrM_+_7006 | 0.12 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr1_+_128637049 | 0.12 |
ENSRNOT00000018639
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr2_+_86996798 | 0.12 |
ENSRNOT00000070821
|
Zfp455
|
zinc finger protein 455 |
| chr6_+_18880737 | 0.12 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr2_+_184600721 | 0.12 |
ENSRNOT00000075823
|
Gatb
|
glutamyl-tRNA amidotransferase subunit B |
| chr16_-_37177033 | 0.12 |
ENSRNOT00000014015
|
Fbxo8
|
F-box protein 8 |
| chr3_+_56125924 | 0.11 |
ENSRNOT00000011380
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr4_+_127164453 | 0.11 |
ENSRNOT00000017889
|
Kbtbd8
|
kelch repeat and BTB domain containing 8 |
| chr3_+_113918629 | 0.11 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr14_-_45859908 | 0.11 |
ENSRNOT00000086994
|
Pgm2
|
phosphoglucomutase 2 |
| chr13_-_47916185 | 0.11 |
ENSRNOT00000087793
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr19_+_24655626 | 0.11 |
ENSRNOT00000086690
|
AC102976.1
|
|
| chr4_-_150520774 | 0.11 |
ENSRNOT00000009095
|
Zfp9
|
zinc finger protein 9 |
| chr13_-_80775230 | 0.11 |
ENSRNOT00000091389
ENSRNOT00000004762 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr3_+_119561290 | 0.11 |
ENSRNOT00000015843
|
Blvra
|
biliverdin reductase A |
| chr9_+_53013413 | 0.11 |
ENSRNOT00000005313
|
Ankar
|
ankyrin and armadillo repeat containing |
| chr11_+_67101714 | 0.11 |
ENSRNOT00000078936
|
Cd86
|
CD86 molecule |
| chr16_+_84465656 | 0.11 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr14_+_7949239 | 0.11 |
ENSRNOT00000044617
|
Ndufb4l1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4-like 1 |
| chr20_+_1889652 | 0.11 |
ENSRNOT00000068508
ENSRNOT00000000996 |
Olr1746
|
olfactory receptor 1746 |
| chrX_-_106607352 | 0.11 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chr3_+_94530586 | 0.11 |
ENSRNOT00000067860
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chrX_+_159081445 | 0.11 |
ENSRNOT00000056688
|
AABR07042542.1
|
|
| chr9_+_73378057 | 0.11 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr1_+_61382118 | 0.11 |
ENSRNOT00000050119
|
Zfp52
|
zinc finger protein 52 |
| chr10_+_84966989 | 0.11 |
ENSRNOT00000013580
|
Scrn2
|
secernin 2 |
| chr1_+_101603222 | 0.11 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr7_+_132378273 | 0.10 |
ENSRNOT00000010990
|
LOC690142
|
hypothetical protein LOC690142 |
| chr1_+_62133816 | 0.10 |
ENSRNOT00000034746
|
LOC102555377
|
zinc finger protein 182-like |
| chr5_+_155934490 | 0.10 |
ENSRNOT00000077933
|
LOC100911993
|
ubiquitin carboxyl-terminal hydrolase 48-like |
| chr14_+_113530470 | 0.10 |
ENSRNOT00000004919
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr1_+_227240383 | 0.10 |
ENSRNOT00000074127
|
Ms4a6e
|
membrane spanning 4-domains A6E |
| chr4_-_162498052 | 0.10 |
ENSRNOT00000066349
|
LOC100360260
|
chromobox homolog 3-like |
| chr2_-_60461458 | 0.10 |
ENSRNOT00000024285
|
Brix1
|
BRX1, biogenesis of ribosomes |
| chr1_+_165237847 | 0.10 |
ENSRNOT00000022963
|
Pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr14_+_17228856 | 0.10 |
ENSRNOT00000003082
|
Cxcl9
|
C-X-C motif chemokine ligand 9 |
| chr5_+_36566783 | 0.10 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chrM_+_7919 | 0.10 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr9_-_38495126 | 0.10 |
ENSRNOT00000016933
ENSRNOT00000090385 |
Rab23
|
RAB23, member RAS oncogene family |
| chr1_-_235405831 | 0.10 |
ENSRNOT00000071578
|
AABR07006458.1
|
|
| chr12_-_1195591 | 0.10 |
ENSRNOT00000001446
|
Stard13
|
StAR-related lipid transfer domain containing 13 |
| chr8_+_79638696 | 0.10 |
ENSRNOT00000085959
|
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 |
| chr2_+_260444227 | 0.10 |
ENSRNOT00000064249
|
Slc44a5
|
solute carrier family 44, member 5 |
| chr7_-_140502441 | 0.10 |
ENSRNOT00000089544
|
Kmt2d
|
lysine methyltransferase 2D |
| chr20_+_34258791 | 0.10 |
ENSRNOT00000000468
|
Slc35f1
|
solute carrier family 35, member F1 |
| chr1_-_67094567 | 0.10 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr15_+_62406873 | 0.10 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr16_+_21399553 | 0.10 |
ENSRNOT00000034446
|
LOC108348302
|
zinc finger protein 709-like |
| chr17_-_86657473 | 0.10 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
| chr4_+_31333970 | 0.09 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr3_-_21669957 | 0.09 |
ENSRNOT00000012354
ENSRNOT00000078718 |
Rc3h2
|
ring finger and CCCH-type domains 2 |
| chr2_+_118910587 | 0.09 |
ENSRNOT00000065518
|
Zfp639
|
zinc finger protein 639 |
| chr8_+_29714285 | 0.09 |
ENSRNOT00000042890
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr2_-_227890077 | 0.09 |
ENSRNOT00000021335
|
Ndst3
|
N-deacetylase and N-sulfotransferase 3 |
| chr1_+_261180007 | 0.09 |
ENSRNOT00000072181
|
Zdhhc16
|
zinc finger, DHHC-type containing 16 |
| chr6_-_108076186 | 0.09 |
ENSRNOT00000014814
|
Fam161b
|
family with sequence similarity 161, member B |
| chr8_-_49158971 | 0.09 |
ENSRNOT00000020573
|
Kmt2a
|
lysine methyltransferase 2A |
| chr17_+_89951752 | 0.09 |
ENSRNOT00000057572
ENSRNOT00000091023 |
Abi1
|
abl-interactor 1 |
| chr12_+_6403940 | 0.09 |
ENSRNOT00000083484
|
B3glct
|
beta 3-glucosyltransferase |
| chr8_-_96516975 | 0.09 |
ENSRNOT00000067996
|
RGD1560775
|
similar to RIKEN cDNA 4930579C12 gene |
| chr2_-_122756842 | 0.09 |
ENSRNOT00000080181
|
Ccdc144b
|
coiled-coil domain containing 144B |
| chr14_-_66978499 | 0.09 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr18_+_86116044 | 0.09 |
ENSRNOT00000058160
ENSRNOT00000076159 |
Rttn
|
rotatin |
| chr9_+_12420368 | 0.09 |
ENSRNOT00000071620
|
AABR07066693.1
|
|
| chr3_-_133131192 | 0.09 |
ENSRNOT00000055630
|
Tasp1
|
taspase 1 |
| chr15_-_28104206 | 0.09 |
ENSRNOT00000032536
|
Ang2
|
angiogenin, ribonuclease A family, member 2 |
| chr7_+_27174882 | 0.09 |
ENSRNOT00000051181
|
Glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr9_+_12475006 | 0.09 |
ENSRNOT00000079703
|
LOC100912293
|
uncharacterized LOC100912293 |
| chr1_-_88516129 | 0.09 |
ENSRNOT00000087232
|
AABR07002893.1
|
|
| chr14_+_108415068 | 0.09 |
ENSRNOT00000083763
|
Pus10
|
pseudouridylate synthase 10 |
| chr11_+_68633160 | 0.09 |
ENSRNOT00000063795
|
Sec22a
|
SEC22 homolog A, vesicle trafficking protein |
| chr17_+_23661429 | 0.09 |
ENSRNOT00000046523
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr10_-_52290657 | 0.09 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr3_-_165700489 | 0.09 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr2_+_72006099 | 0.09 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr12_+_19196611 | 0.09 |
ENSRNOT00000001801
|
Azgp1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr5_+_25253010 | 0.09 |
ENSRNOT00000061333
|
Rbm12b
|
RNA binding motif protein 12B |
| chr19_+_15081590 | 0.08 |
ENSRNOT00000024187
|
Ces1f
|
carboxylesterase 1F |
| chr2_-_219262901 | 0.08 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr4_+_162281574 | 0.08 |
ENSRNOT00000087850
|
Clec2d
|
C-type lectin domain family 2, member D |
| chr1_+_99505677 | 0.08 |
ENSRNOT00000024645
|
Zfp719
|
zinc finger protein 719 |
| chr8_-_12355091 | 0.08 |
ENSRNOT00000009318
|
Cep57
|
centrosomal protein 57 |
| chr9_-_1012450 | 0.08 |
ENSRNOT00000051449
|
LOC100359951
|
ribosomal protein S20-like |
| chr4_-_176909075 | 0.08 |
ENSRNOT00000067489
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chr2_-_40386669 | 0.08 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr4_+_112662609 | 0.08 |
ENSRNOT00000009104
|
Gcfc2
|
GC-rich sequence DNA-binding factor 2 |
| chr1_+_164706276 | 0.08 |
ENSRNOT00000024311
|
Olr36
|
olfactory receptor 36 |
| chr5_-_164648328 | 0.08 |
ENSRNOT00000090811
|
Zfp933
|
zinc finger protein 933 |
| chrX_-_70978952 | 0.08 |
ENSRNOT00000076910
|
Slc7a3
|
solute carrier family 7 member 3 |
| chr19_-_41567013 | 0.08 |
ENSRNOT00000022757
|
Zfp612
|
zinc finger protein 612 |
| chr10_-_87521514 | 0.08 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr3_+_21764377 | 0.08 |
ENSRNOT00000063938
|
Gpr21
|
G protein-coupled receptor 21 |
| chr14_-_18704059 | 0.08 |
ENSRNOT00000081455
|
Mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr12_-_21362205 | 0.08 |
ENSRNOT00000064787
|
LOC108348155
|
paired immunoglobulin-like type 2 receptor beta-2 |
| chr5_-_73552798 | 0.08 |
ENSRNOT00000022836
|
Ikbkap
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Lhx2_Hoxc5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071623 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.1 | 0.3 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.3 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.1 | 0.3 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.1 | 0.3 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.1 | 0.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 0.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.3 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.2 | GO:0000962 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) |
| 0.1 | 0.2 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.2 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.2 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.1 | GO:0072702 | myofibroblast differentiation(GO:0036446) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.5 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:2000458 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.0 | 0.1 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.0 | 0.1 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:2000688 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 0.0 | 0.1 | GO:0035722 | monocyte extravasation(GO:0035696) interleukin-12-mediated signaling pathway(GO:0035722) interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-12(GO:0071349) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0006431 | methionyl-tRNA aminoacylation(GO:0006431) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.0 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.2 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 1.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.1 | GO:0007309 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0045963 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.0 | GO:0048382 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) mesendoderm development(GO:0048382) regulation of cardiac ventricle development(GO:1904412) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.0 | GO:0036334 | epidermal stem cell homeostasis(GO:0036334) |
| 0.0 | 0.0 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.0 | GO:0043179 | rhythmic excitation(GO:0043179) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.0 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.0 | GO:0010034 | response to acetate(GO:0010034) |
| 0.0 | 0.0 | GO:0042125 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0034035 | flavonoid metabolic process(GO:0009812) purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.0 | GO:0060383 | positive regulation of DNA strand elongation(GO:0060383) |
| 0.0 | 0.1 | GO:0002911 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.1 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.0 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.4 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.0 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.0 | 0.0 | GO:0021629 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.0 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.0 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.0 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.0 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.0 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0044317 | rod spherule(GO:0044317) |
| 0.1 | 0.2 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.1 | 0.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 0.2 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 1.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) Ndc80 complex(GO:0031262) |
| 0.0 | 0.0 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 1.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0070401 | NADP-retinol dehydrogenase activity(GO:0052650) NADP+ binding(GO:0070401) |
| 0.0 | 0.1 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.2 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0004825 | methionine-tRNA ligase activity(GO:0004825) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.0 | GO:0098603 | selenol Se-methyltransferase activity(GO:0098603) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.0 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.0 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.0 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.0 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |