Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Lef1
Z-value: 0.39
Transcription factors associated with Lef1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lef1
|
ENSRNOG00000010121 | lymphoid enhancer binding factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lef1 | rn6_v1_chr2_+_236233239_236233239 | 0.55 | 3.3e-01 | Click! |
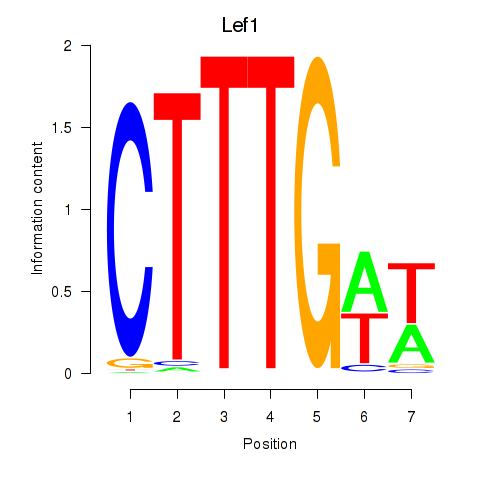
Activity profile of Lef1 motif
Sorted Z-values of Lef1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_28515025 | 0.17 |
ENSRNOT00000088033
ENSRNOT00000005317 ENSRNOT00000081141 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr3_+_147585947 | 0.16 |
ENSRNOT00000006833
|
Scrt2
|
scratch family transcriptional repressor 2 |
| chr16_-_74264142 | 0.16 |
ENSRNOT00000026067
|
Dkk4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr7_-_93826665 | 0.16 |
ENSRNOT00000011344
|
Tnfrsf11b
|
TNF receptor superfamily member 11B |
| chr2_+_150756185 | 0.14 |
ENSRNOT00000088461
ENSRNOT00000036808 |
Mbnl1
|
muscleblind-like splicing regulator 1 |
| chr10_-_27179254 | 0.13 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr2_-_142686577 | 0.13 |
ENSRNOT00000014562
|
Nhlrc3
|
NHL repeat containing 3 |
| chr3_+_47677720 | 0.12 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr11_+_9642365 | 0.12 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr7_-_98098268 | 0.12 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chr3_+_131351587 | 0.11 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr18_+_30900291 | 0.11 |
ENSRNOT00000060461
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7 |
| chr10_-_83128297 | 0.11 |
ENSRNOT00000082160
|
Kat7
|
lysine acetyltransferase 7 |
| chr5_-_168734296 | 0.11 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr3_+_59981959 | 0.11 |
ENSRNOT00000030460
|
Sp9
|
Sp9 transcription factor |
| chr8_+_49418965 | 0.11 |
ENSRNOT00000021819
|
Scn2b
|
sodium voltage-gated channel beta subunit 2 |
| chr9_+_14551758 | 0.10 |
ENSRNOT00000017157
|
Nfya
|
nuclear transcription factor Y subunit alpha |
| chr14_+_2613406 | 0.10 |
ENSRNOT00000000083
|
Tmed5
|
transmembrane p24 trafficking protein 5 |
| chr2_-_140618405 | 0.09 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr3_+_148541909 | 0.09 |
ENSRNOT00000012187
|
Ccm2l
|
CCM2 like scaffolding protein |
| chr20_-_27208041 | 0.09 |
ENSRNOT00000084720
|
Rufy2
|
RUN and FYVE domain containing 2 |
| chr13_+_34400170 | 0.09 |
ENSRNOT00000061516
ENSRNOT00000061515 ENSRNOT00000061513 ENSRNOT00000084506 ENSRNOT00000086641 |
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr5_-_79222687 | 0.09 |
ENSRNOT00000010516
|
Akna
|
AT-hook transcription factor |
| chr18_+_27424328 | 0.08 |
ENSRNOT00000033784
|
Kif20a
|
kinesin family member 20A |
| chr2_+_205783252 | 0.08 |
ENSRNOT00000025633
ENSRNOT00000025600 |
Trim33
|
tripartite motif-containing 33 |
| chr11_-_62451149 | 0.08 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_+_142453013 | 0.08 |
ENSRNOT00000056573
|
AABR07061755.1
|
|
| chr1_+_151439409 | 0.08 |
ENSRNOT00000022060
ENSRNOT00000050639 |
Grm5
|
glutamate metabotropic receptor 5 |
| chrX_-_26376467 | 0.08 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr2_+_238257031 | 0.07 |
ENSRNOT00000016066
|
Ints12
|
integrator complex subunit 12 |
| chr2_+_179952227 | 0.07 |
ENSRNOT00000015081
|
Pdgfc
|
platelet derived growth factor C |
| chr2_-_165739874 | 0.07 |
ENSRNOT00000014384
|
Kpna4
|
karyopherin subunit alpha 4 |
| chr18_+_14756684 | 0.07 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr13_+_52976507 | 0.07 |
ENSRNOT00000090599
ENSRNOT00000011324 |
Kif21b
|
kinesin family member 21B |
| chr15_-_2966576 | 0.07 |
ENSRNOT00000070893
ENSRNOT00000017383 |
Kat6b
|
lysine acetyltransferase 6B |
| chr17_+_13670520 | 0.07 |
ENSRNOT00000019442
|
Shc3
|
SHC adaptor protein 3 |
| chr3_+_116899878 | 0.07 |
ENSRNOT00000090802
ENSRNOT00000066101 |
Sema6d
|
semaphorin 6D |
| chr18_+_30909490 | 0.07 |
ENSRNOT00000026967
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8 |
| chr1_-_131460473 | 0.07 |
ENSRNOT00000084336
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_+_142686724 | 0.07 |
ENSRNOT00000014614
|
Proser1
|
proline and serine rich 1 |
| chr6_+_147876237 | 0.06 |
ENSRNOT00000056649
|
Tmem196
|
transmembrane protein 196 |
| chrX_+_156655960 | 0.06 |
ENSRNOT00000085723
|
Mecp2
|
methyl CpG binding protein 2 |
| chr7_-_36408588 | 0.06 |
ENSRNOT00000063946
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr8_+_118821729 | 0.06 |
ENSRNOT00000028409
|
Setd2
|
SET domain containing 2 |
| chr14_+_63095720 | 0.06 |
ENSRNOT00000006071
|
Ppargc1a
|
PPARG coactivator 1 alpha |
| chr2_-_147693082 | 0.06 |
ENSRNOT00000022507
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr1_-_198316882 | 0.06 |
ENSRNOT00000085304
ENSRNOT00000064985 |
Taok2
|
TAO kinase 2 |
| chr8_-_78397123 | 0.06 |
ENSRNOT00000087270
ENSRNOT00000084925 |
Tcf12
|
transcription factor 12 |
| chr20_-_31597830 | 0.06 |
ENSRNOT00000085877
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr10_-_27179900 | 0.06 |
ENSRNOT00000082445
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr6_+_99625306 | 0.06 |
ENSRNOT00000008573
|
Plekhg3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr3_-_43119159 | 0.06 |
ENSRNOT00000041394
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr15_+_37171052 | 0.06 |
ENSRNOT00000011684
|
Zmym2
|
zinc finger MYM-type containing 2 |
| chr8_-_73164620 | 0.06 |
ENSRNOT00000031988
|
Tln2
|
talin 2 |
| chr8_-_84506328 | 0.06 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr9_+_2202511 | 0.06 |
ENSRNOT00000017556
|
Satb1
|
SATB homeobox 1 |
| chr10_+_81913689 | 0.06 |
ENSRNOT00000003780
|
Tob1
|
transducer of ErbB-2.1 |
| chr5_+_4373626 | 0.06 |
ENSRNOT00000064946
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr2_+_205553163 | 0.06 |
ENSRNOT00000039572
|
Nras
|
neuroblastoma RAS viral oncogene homolog |
| chr12_+_27155587 | 0.05 |
ENSRNOT00000044800
|
AABR07035916.1
|
|
| chr10_-_85574889 | 0.05 |
ENSRNOT00000072274
|
LOC691153
|
hypothetical protein LOC691153 |
| chr8_-_36467627 | 0.05 |
ENSRNOT00000082346
|
Fam118b
|
family with sequence similarity 118, member B |
| chr13_+_71110064 | 0.05 |
ENSRNOT00000088329
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr3_+_56766475 | 0.05 |
ENSRNOT00000078819
|
Sp5
|
Sp5 transcription factor |
| chr6_-_147172022 | 0.05 |
ENSRNOT00000080675
|
Itgb8
|
integrin subunit beta 8 |
| chr13_-_73055631 | 0.05 |
ENSRNOT00000081892
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr7_-_140546908 | 0.05 |
ENSRNOT00000077502
|
Kmt2d
|
lysine methyltransferase 2D |
| chr9_+_19451630 | 0.05 |
ENSRNOT00000065048
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chrX_-_115175299 | 0.05 |
ENSRNOT00000074322
|
Dcx
|
doublecortin |
| chr13_-_51076852 | 0.04 |
ENSRNOT00000078993
|
Adora1
|
adenosine A1 receptor |
| chr9_+_49479023 | 0.04 |
ENSRNOT00000050922
ENSRNOT00000077111 |
Pou3f3
|
POU class 3 homeobox 3 |
| chr8_+_117620317 | 0.04 |
ENSRNOT00000084220
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr15_+_105851542 | 0.04 |
ENSRNOT00000086959
|
Rap2a
|
RAS related protein 2a |
| chr12_-_47793534 | 0.04 |
ENSRNOT00000001588
|
Fam222a
|
family with sequence similarity 222, member A |
| chr5_+_122100099 | 0.04 |
ENSRNOT00000007738
|
Pde4b
|
phosphodiesterase 4B |
| chr17_-_51912496 | 0.04 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr2_+_220432037 | 0.04 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr8_+_63600663 | 0.04 |
ENSRNOT00000012644
|
Hcn4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr8_+_59457018 | 0.04 |
ENSRNOT00000017900
|
Ireb2
|
iron responsive element binding protein 2 |
| chr1_-_6970040 | 0.04 |
ENSRNOT00000016273
|
Utrn
|
utrophin |
| chr20_-_31598118 | 0.04 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr17_+_23135985 | 0.04 |
ENSRNOT00000090794
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr19_+_39543242 | 0.04 |
ENSRNOT00000018837
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr10_+_88987558 | 0.04 |
ENSRNOT00000026878
|
Hsd17b1
|
hydroxysteroid (17-beta) dehydrogenase 1 |
| chr2_+_196013799 | 0.04 |
ENSRNOT00000084023
|
Pogz
|
pogo transposable element with ZNF domain |
| chr17_+_82065937 | 0.04 |
ENSRNOT00000051349
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chr13_+_98924962 | 0.04 |
ENSRNOT00000074148
|
Lin9
|
lin-9 DREAM MuvB core complex component |
| chr5_+_128083118 | 0.04 |
ENSRNOT00000079161
|
Zcchc11
|
zinc finger CCHC-type containing 11 |
| chr2_-_220535751 | 0.03 |
ENSRNOT00000089082
|
Palmd
|
palmdelphin |
| chr15_+_3996048 | 0.03 |
ENSRNOT00000078653
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr17_-_84830185 | 0.03 |
ENSRNOT00000040697
|
Skida1
|
SKI/DACH domain containing 1 |
| chr5_-_144479306 | 0.03 |
ENSRNOT00000087697
|
Ago1
|
argonaute 1, RISC catalytic component |
| chr13_-_52514875 | 0.03 |
ENSRNOT00000064758
|
Nav1
|
neuron navigator 1 |
| chr2_-_211001258 | 0.03 |
ENSRNOT00000037336
|
Atxn7l2
|
ataxin 7-like 2 |
| chr1_-_134870255 | 0.03 |
ENSRNOT00000055829
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr9_-_38196273 | 0.03 |
ENSRNOT00000044452
|
Dst
|
dystonin |
| chr13_-_72063347 | 0.03 |
ENSRNOT00000090544
ENSRNOT00000003869 |
Cacna1e
|
calcium voltage-gated channel subunit alpha1 E |
| chr11_-_31213787 | 0.03 |
ENSRNOT00000002800
|
Paxbp1
|
PAX3 and PAX7 binding protein 1 |
| chr13_-_81214494 | 0.03 |
ENSRNOT00000004950
ENSRNOT00000082385 |
Prrx1
|
paired related homeobox 1 |
| chr10_-_74376270 | 0.03 |
ENSRNOT00000079311
|
Gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr13_-_84619023 | 0.03 |
ENSRNOT00000089827
ENSRNOT00000045056 |
Pogk
|
pogo transposable element with KRAB domain |
| chr15_+_86243148 | 0.03 |
ENSRNOT00000084471
ENSRNOT00000090727 |
Lmo7
|
LIM domain 7 |
| chr16_-_71203609 | 0.03 |
ENSRNOT00000088458
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr5_+_25168295 | 0.03 |
ENSRNOT00000020245
|
Fsbp
|
fibrinogen silencer binding protein |
| chr6_-_72596446 | 0.03 |
ENSRNOT00000008459
|
Hectd1
|
HECT domain E3 ubiquitin protein ligase 1 |
| chr11_-_35697072 | 0.03 |
ENSRNOT00000039999
|
Erg
|
ERG, ETS transcription factor |
| chr5_-_139748489 | 0.03 |
ENSRNOT00000078741
|
Nfyc
|
nuclear transcription factor Y subunit gamma |
| chr1_-_218100272 | 0.03 |
ENSRNOT00000028411
ENSRNOT00000088588 |
Ccnd1
|
cyclin D1 |
| chr3_+_58164931 | 0.03 |
ENSRNOT00000002078
|
Dlx1
|
distal-less homeobox 1 |
| chr13_-_111581018 | 0.03 |
ENSRNOT00000083072
ENSRNOT00000077981 |
Sertad4
|
SERTA domain containing 4 |
| chr2_+_144861455 | 0.02 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr1_-_221015929 | 0.02 |
ENSRNOT00000028137
|
Sipa1
|
signal-induced proliferation-associated 1 |
| chr8_-_14880644 | 0.02 |
ENSRNOT00000015977
|
Fat3
|
FAT atypical cadherin 3 |
| chr8_-_96132634 | 0.02 |
ENSRNOT00000041655
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr3_+_149741312 | 0.02 |
ENSRNOT00000070854
|
LOC100911637
|
high mobility group protein B1-like |
| chr8_+_111210811 | 0.02 |
ENSRNOT00000011347
|
Amotl2
|
angiomotin like 2 |
| chr14_+_66598259 | 0.02 |
ENSRNOT00000049743
|
Kcnip4
|
potassium voltage-gated channel interacting protein 4 |
| chrX_-_32095867 | 0.02 |
ENSRNOT00000049947
ENSRNOT00000080730 |
Ace2
|
angiotensin I converting enzyme 2 |
| chr10_-_86004096 | 0.02 |
ENSRNOT00000091978
ENSRNOT00000066855 |
Stac2
|
SH3 and cysteine rich domain 2 |
| chr2_-_219458271 | 0.02 |
ENSRNOT00000064735
|
Cdc14a
|
cell division cycle 14A |
| chr7_-_132984110 | 0.02 |
ENSRNOT00000046626
|
Hmgb1-ps3
|
high mobility group box 1, pseudogene 3 |
| chr15_-_41338284 | 0.02 |
ENSRNOT00000077225
|
Tnfrsf19
|
TNF receptor superfamily member 19 |
| chr18_-_27424090 | 0.02 |
ENSRNOT00000087968
|
Brd8
|
bromodomain containing 8 |
| chr9_-_61810417 | 0.02 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr14_-_35652709 | 0.02 |
ENSRNOT00000003080
|
Gsx2
|
GS homeobox 2 |
| chr3_-_51643140 | 0.02 |
ENSRNOT00000006646
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chrX_+_9436707 | 0.02 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr13_-_91872954 | 0.02 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr8_+_118206698 | 0.02 |
ENSRNOT00000082607
ENSRNOT00000064140 |
Smarcc1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chrX_+_86126157 | 0.02 |
ENSRNOT00000006992
|
Klhl4
|
kelch-like family member 4 |
| chr8_-_118182559 | 0.02 |
ENSRNOT00000084838
|
Dhx30
|
DExH-box helicase 30 |
| chrX_-_4945944 | 0.02 |
ENSRNOT00000077238
|
Kdm6a
|
lysine demethylase 6A |
| chr18_+_79773608 | 0.02 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr15_-_100303807 | 0.02 |
ENSRNOT00000041344
|
LOC102548369
|
40S ribosomal protein S16-like |
| chr5_-_7874909 | 0.02 |
ENSRNOT00000064774
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr9_-_81772851 | 0.02 |
ENSRNOT00000032719
|
Usp37
|
ubiquitin specific peptidase 37 |
| chr10_+_94407559 | 0.02 |
ENSRNOT00000013046
|
Ddx42
|
DEAD-box helicase 42 |
| chr10_-_89700283 | 0.01 |
ENSRNOT00000028213
|
Etv4
|
ets variant 4 |
| chr9_+_65279675 | 0.01 |
ENSRNOT00000019486
|
Bzw1
|
basic leucine zipper and W2 domains 1 |
| chr9_+_46657575 | 0.01 |
ENSRNOT00000083322
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr6_-_147172813 | 0.01 |
ENSRNOT00000066545
|
Itgb8
|
integrin subunit beta 8 |
| chr14_-_2474780 | 0.01 |
ENSRNOT00000074874
|
LOC100910207
|
protein Dr1-like |
| chr6_+_73358112 | 0.01 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr6_-_22092346 | 0.01 |
ENSRNOT00000083128
ENSRNOT00000006830 |
Birc6
|
baculoviral IAP repeat-containing 6 |
| chr4_+_78240385 | 0.01 |
ENSRNOT00000011041
|
Zfp775
|
zinc finger protein 775 |
| chr2_-_188660179 | 0.01 |
ENSRNOT00000027935
|
Efna4
|
ephrin A4 |
| chr3_-_107760550 | 0.01 |
ENSRNOT00000077091
ENSRNOT00000051638 |
Meis2
|
Meis homeobox 2 |
| chr13_-_95250235 | 0.01 |
ENSRNOT00000085648
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr8_+_70630767 | 0.01 |
ENSRNOT00000051353
|
Igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr4_+_179481263 | 0.01 |
ENSRNOT00000021284
|
Etfrf1
|
electron transfer flavoprotein regulatory factor 1 |
| chr2_-_57935334 | 0.01 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr6_+_108820774 | 0.01 |
ENSRNOT00000089683
ENSRNOT00000080038 ENSRNOT00000006492 |
Ylpm1
|
YLP motif containing 1 |
| chr8_-_64268555 | 0.01 |
ENSRNOT00000013581
ENSRNOT00000084758 |
Arih1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr6_+_147876557 | 0.01 |
ENSRNOT00000080090
|
Tmem196
|
transmembrane protein 196 |
| chr5_+_126976456 | 0.01 |
ENSRNOT00000014275
|
Ndc1
|
NDC1 transmembrane nucleoporin |
| chr6_-_107080524 | 0.01 |
ENSRNOT00000011662
|
Zfyve1
|
zinc finger FYVE-type containing 1 |
| chr2_-_139528162 | 0.01 |
ENSRNOT00000014317
|
Slc7a11
|
solute carrier family 7 member 11 |
| chr18_-_58097316 | 0.01 |
ENSRNOT00000088527
|
AABR07032265.1
|
|
| chr9_-_4447715 | 0.01 |
ENSRNOT00000061882
|
Kat2b
|
lysine acetyltransferase 2B |
| chr18_+_46148849 | 0.01 |
ENSRNOT00000026724
|
Prr16
|
proline rich 16 |
| chrX_-_18726732 | 0.00 |
ENSRNOT00000026214
|
Ubqln2
|
ubiquilin 2 |
| chr6_+_145546595 | 0.00 |
ENSRNOT00000007112
|
Rapgef5
|
Rap guanine nucleotide exchange factor 5 |
| chr10_+_89285855 | 0.00 |
ENSRNOT00000028033
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr14_-_44845218 | 0.00 |
ENSRNOT00000004003
|
Klhl5
|
kelch-like family member 5 |
| chr14_-_2700977 | 0.00 |
ENSRNOT00000000086
|
Mtf2
|
metal response element binding transcription factor 2 |
| chr3_+_8643936 | 0.00 |
ENSRNOT00000077797
|
Set
|
SET nuclear proto-oncogene |
| chr10_-_87541851 | 0.00 |
ENSRNOT00000089610
|
RGD1561684
|
similar to keratin associated protein 2-4 |
| chrX_+_54266687 | 0.00 |
ENSRNOT00000058309
|
LOC100363193
|
LRRGT00076-like |
| chr9_-_111213098 | 0.00 |
ENSRNOT00000087355
|
Gin1
|
gypsy retrotransposon integrase 1 |
| chr19_-_29016521 | 0.00 |
ENSRNOT00000049860
|
LOC685160
|
similar to spermatogenesis associated glutamate (E)-rich protein 4d |
| chr20_+_5262120 | 0.00 |
ENSRNOT00000000535
|
Brd2
|
bromodomain containing 2 |
| chr20_-_32258286 | 0.00 |
ENSRNOT00000082114
|
Ddx50
|
DExD-box helicase 50 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Lef1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 | 0.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.1 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:0060849 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.1 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:1900453 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) negative regulation of long term synaptic depression(GO:1900453) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.0 | GO:0072021 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |