Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
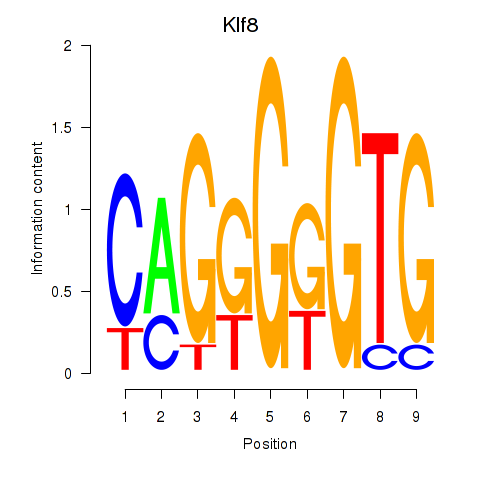
Results for Klf8
Z-value: 0.26
Transcription factors associated with Klf8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf8
|
ENSRNOG00000039057 | Kruppel-like factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf8 | rn6_v1_chrX_-_18890090_18890090 | -0.05 | 9.4e-01 | Click! |
Activity profile of Klf8 motif
Sorted Z-values of Klf8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_15301998 | 0.14 |
ENSRNOT00000016400
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr10_-_15590220 | 0.10 |
ENSRNOT00000048977
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr1_-_84812486 | 0.08 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chrX_+_15273933 | 0.07 |
ENSRNOT00000075082
|
LOC108348091
|
erythroid transcription factor |
| chrX_-_38196060 | 0.06 |
ENSRNOT00000006741
ENSRNOT00000006438 |
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr2_-_46544457 | 0.06 |
ENSRNOT00000015680
|
Fst
|
follistatin |
| chr3_-_5693147 | 0.05 |
ENSRNOT00000008660
|
Fam163b
|
family with sequence similarity 163, member B |
| chr4_-_100883275 | 0.05 |
ENSRNOT00000022846
|
LOC100364435
|
thymosin, beta 10-like |
| chr20_+_4576514 | 0.05 |
ENSRNOT00000090125
ENSRNOT00000047370 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr11_+_32211115 | 0.05 |
ENSRNOT00000087452
|
Mrps6
|
mitochondrial ribosomal protein S6 |
| chr7_-_121311565 | 0.04 |
ENSRNOT00000092252
|
Rpl3
|
ribosomal protein L3 |
| chr3_+_110466790 | 0.04 |
ENSRNOT00000089434
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr12_+_23544287 | 0.04 |
ENSRNOT00000001938
|
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr20_+_4576057 | 0.04 |
ENSRNOT00000081456
ENSRNOT00000085701 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr7_+_142877086 | 0.04 |
ENSRNOT00000088730
|
Grasp
|
general receptor for phosphoinositides 1 associated scaffold protein |
| chr14_-_36554580 | 0.04 |
ENSRNOT00000002869
|
Rasl11b
|
RAS-like family 11 member B |
| chr2_+_251817694 | 0.03 |
ENSRNOT00000019964
|
RGD1560065
|
similar to RIKEN cDNA 2410004B18 |
| chr10_-_48860902 | 0.03 |
ENSRNOT00000004127
|
Cenpv
|
centromere protein V |
| chr18_+_47740328 | 0.03 |
ENSRNOT00000025119
|
Sncaip
|
synuclein, alpha interacting protein |
| chr2_-_192697800 | 0.03 |
ENSRNOT00000066614
|
NEWGENE_2324572
|
small proline rich protein 4 |
| chr13_-_53108713 | 0.03 |
ENSRNOT00000035404
|
RGD1311892
|
similar to hypothetical protein FLJ10901 |
| chr10_-_29450644 | 0.03 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr16_+_9563218 | 0.03 |
ENSRNOT00000035915
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr6_+_129538982 | 0.03 |
ENSRNOT00000083626
ENSRNOT00000082522 |
Ak7
|
adenylate kinase 7 |
| chr12_-_22478752 | 0.03 |
ENSRNOT00000089392
ENSRNOT00000086915 |
Ache
|
acetylcholinesterase |
| chr2_+_231962517 | 0.03 |
ENSRNOT00000014570
|
Neurog2
|
neurogenin 2 |
| chr11_-_87236445 | 0.03 |
ENSRNOT00000057806
|
Tssk2
|
testis-specific serine kinase 2 |
| chr20_-_4943564 | 0.03 |
ENSRNOT00000049310
|
RT1-CE1
|
RT1 class I, locus1 |
| chr10_+_109107389 | 0.03 |
ENSRNOT00000068437
ENSRNOT00000005687 |
Baiap2
|
BAI1-associated protein 2 |
| chr10_-_90999506 | 0.03 |
ENSRNOT00000034401
|
Gfap
|
glial fibrillary acidic protein |
| chr1_-_226657408 | 0.02 |
ENSRNOT00000028102
|
Tkfc
|
triokinase and FMN cyclase |
| chr19_-_59502161 | 0.02 |
ENSRNOT00000075292
|
Irf2bp2
|
interferon regulatory factor 2 binding protein 2 |
| chr20_-_28814636 | 0.02 |
ENSRNOT00000086030
|
Sept10
|
septin 10 |
| chr1_-_80271001 | 0.02 |
ENSRNOT00000034266
|
Cd3eap
|
CD3e molecule associated protein |
| chr9_+_81672758 | 0.02 |
ENSRNOT00000020646
|
Ctdsp1
|
CTD small phosphatase 1 |
| chr10_+_10644572 | 0.02 |
ENSRNOT00000004026
|
Ppl
|
periplakin |
| chr3_+_175982777 | 0.02 |
ENSRNOT00000082073
|
Ntsr1
|
neurotensin receptor 1 |
| chr2_+_23385183 | 0.02 |
ENSRNOT00000014860
|
Arsb
|
arylsulfatase B |
| chr2_+_205160405 | 0.02 |
ENSRNOT00000035605
|
Tspan2
|
tetraspanin 2 |
| chr3_+_100769839 | 0.02 |
ENSRNOT00000077703
|
Bdnf
|
brain-derived neurotrophic factor |
| chr1_-_189911571 | 0.02 |
ENSRNOT00000080996
ENSRNOT00000088536 |
Zp2
|
zona pellucida glycoprotein 2 |
| chr3_+_2158995 | 0.02 |
ENSRNOT00000010270
|
Zmynd19
|
zinc finger, MYND-type containing 19 |
| chr8_+_116054465 | 0.02 |
ENSRNOT00000040056
|
Cish
|
cytokine inducible SH2-containing protein |
| chr1_-_247169693 | 0.02 |
ENSRNOT00000079875
|
Ak3
|
adenylate kinase 3 |
| chr3_+_113976687 | 0.02 |
ENSRNOT00000022437
|
Eif3j
|
eukaryotic translation initiation factor 3, subunit J |
| chr18_+_70427007 | 0.02 |
ENSRNOT00000087959
ENSRNOT00000019512 |
Myo5b
|
myosin Vb |
| chr9_-_82699551 | 0.02 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chr4_-_146839397 | 0.02 |
ENSRNOT00000010338
|
Vgll4
|
vestigial-like family member 4 |
| chr7_+_145068286 | 0.02 |
ENSRNOT00000088956
ENSRNOT00000065753 |
Nckap1l
|
NCK associated protein 1 like |
| chr1_-_89488223 | 0.02 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr2_-_84437084 | 0.02 |
ENSRNOT00000077598
|
Ankrd33b
|
ankyrin repeat domain 33B |
| chr14_-_78377825 | 0.02 |
ENSRNOT00000068104
|
AABR07015812.1
|
|
| chr17_-_35037076 | 0.02 |
ENSRNOT00000090946
|
Dusp22
|
dual specificity phosphatase 22 |
| chr9_+_99998275 | 0.02 |
ENSRNOT00000074395
|
Gpc1
|
glypican 1 |
| chr1_-_167347662 | 0.02 |
ENSRNOT00000027641
ENSRNOT00000076592 |
Rhog
|
ras homolog family member G |
| chr1_-_242083484 | 0.02 |
ENSRNOT00000065921
|
Tjp2
|
tight junction protein 2 |
| chr6_-_38228379 | 0.01 |
ENSRNOT00000084924
|
Mycn
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr10_+_69434941 | 0.01 |
ENSRNOT00000009756
|
Ccl11
|
C-C motif chemokine ligand 11 |
| chr2_+_85377318 | 0.01 |
ENSRNOT00000016506
ENSRNOT00000085094 |
Sema5a
|
semaphorin 5A |
| chr1_-_229601032 | 0.01 |
ENSRNOT00000016690
|
Cntf
|
ciliary neurotrophic factor |
| chr1_+_84304228 | 0.01 |
ENSRNOT00000024771
|
Prx
|
periaxin |
| chr19_+_37127508 | 0.01 |
ENSRNOT00000019656
|
Cbfb
|
core-binding factor, beta subunit |
| chr20_+_11863082 | 0.01 |
ENSRNOT00000001640
|
Fam207a
|
family with sequence similarity 207, member A |
| chr10_+_55169282 | 0.01 |
ENSRNOT00000005423
|
Ccdc42
|
coiled-coil domain containing 42 |
| chr11_-_34598102 | 0.01 |
ENSRNOT00000068743
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr13_+_79378733 | 0.01 |
ENSRNOT00000039221
|
Tnfsf18
|
tumor necrosis factor superfamily member 18 |
| chr16_+_10554136 | 0.01 |
ENSRNOT00000079030
|
Syt15
|
synaptotagmin 15 |
| chr11_-_84047497 | 0.01 |
ENSRNOT00000088821
ENSRNOT00000058092 |
Ap2m1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr15_+_34431357 | 0.01 |
ENSRNOT00000027604
|
Nop9
|
NOP9 nucleolar protein |
| chrX_-_142248369 | 0.01 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr10_+_56237755 | 0.01 |
ENSRNOT00000016163
|
Fxr2
|
FMR1 autosomal homolog 2 |
| chr1_-_53087474 | 0.01 |
ENSRNOT00000017302
|
Ccr6
|
C-C motif chemokine receptor 6 |
| chrM_-_14061 | 0.01 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr4_-_45332420 | 0.01 |
ENSRNOT00000083039
|
Wnt2
|
wingless-type MMTV integration site family member 2 |
| chr6_-_102353403 | 0.01 |
ENSRNOT00000090407
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr8_+_32018560 | 0.01 |
ENSRNOT00000007358
|
Adamts8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr14_+_19319299 | 0.01 |
ENSRNOT00000086542
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr3_-_176706896 | 0.01 |
ENSRNOT00000017337
|
Ptk6
|
protein tyrosine kinase 6 |
| chr18_+_48132414 | 0.01 |
ENSRNOT00000050631
|
Snx2
|
sorting nexin 2 |
| chr7_+_92234597 | 0.01 |
ENSRNOT00000067491
ENSRNOT00000089991 |
Med30
|
mediator complex subunit 30 |
| chr20_+_27129291 | 0.01 |
ENSRNOT00000031053
|
AABR07044933.1
|
|
| chr9_+_93080615 | 0.01 |
ENSRNOT00000024306
|
Psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr2_-_24923128 | 0.01 |
ENSRNOT00000044087
|
Pde8b
|
phosphodiesterase 8B |
| chr1_+_221872220 | 0.01 |
ENSRNOT00000028651
|
Nrxn2
|
neurexin 2 |
| chr5_-_139933764 | 0.01 |
ENSRNOT00000015278
|
LOC680700
|
similar to ribosomal protein L10a |
| chr1_+_192025710 | 0.01 |
ENSRNOT00000077457
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr1_-_84168044 | 0.01 |
ENSRNOT00000084586
ENSRNOT00000028348 |
Shkbp1
|
Sh3kbp1 binding protein 1 |
| chr5_-_168004724 | 0.01 |
ENSRNOT00000024711
|
Park7
|
Parkinsonism associated deglycase |
| chr9_+_113699170 | 0.01 |
ENSRNOT00000017915
|
Twsg1
|
twisted gastrulation BMP signaling modulator 1 |
| chr6_-_76079664 | 0.01 |
ENSRNOT00000033533
|
Ppp2r3c
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr1_-_73753128 | 0.01 |
ENSRNOT00000068459
|
Ttyh1
|
tweety family member 1 |
| chr15_+_24267323 | 0.01 |
ENSRNOT00000015778
|
Fbxo34
|
F-box protein 34 |
| chr8_-_117797427 | 0.01 |
ENSRNOT00000033719
|
Trex1
|
three prime repair exonuclease 1 |
| chr1_-_4653210 | 0.01 |
ENSRNOT00000019121
|
Rab32
|
RAB32, member RAS oncogene family |
| chr1_-_274106502 | 0.01 |
ENSRNOT00000020539
|
Smndc1
|
survival motor neuron domain containing 1 |
| chr1_-_225014538 | 0.01 |
ENSRNOT00000026376
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr8_-_123371257 | 0.01 |
ENSRNOT00000017243
|
Stt3b
|
STT3B, catalytic subunit of the oligosaccharyltransferase complex |
| chr7_+_10937599 | 0.01 |
ENSRNOT00000008758
|
Sirt6
|
sirtuin 6 |
| chr5_+_59348639 | 0.01 |
ENSRNOT00000084031
ENSRNOT00000060264 |
Reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chrX_-_33665821 | 0.01 |
ENSRNOT00000066676
|
Rbbp7
|
RB binding protein 7, chromatin remodeling factor |
| chr2_-_252359798 | 0.01 |
ENSRNOT00000021063
|
Spata1
|
spermatogenesis associated 1 |
| chr7_-_105592804 | 0.01 |
ENSRNOT00000006789
|
Adcy8
|
adenylate cyclase 8 |
| chr12_-_6956914 | 0.01 |
ENSRNOT00000072129
ENSRNOT00000001210 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr2_-_192027225 | 0.01 |
ENSRNOT00000016430
|
Prr9
|
proline rich 9 |
| chr18_+_28594892 | 0.01 |
ENSRNOT00000067702
|
Ube2d2
|
ubiquitin-conjugating enzyme E2D 2 |
| chr6_-_76079283 | 0.01 |
ENSRNOT00000082198
|
Ppp2r3c
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr8_-_67232298 | 0.01 |
ENSRNOT00000049441
|
Spesp1
|
sperm equatorial segment protein 1 |
| chr12_-_47588335 | 0.00 |
ENSRNOT00000001595
|
Ankrd13a
|
ankyrin repeat domain 13a |
| chr10_-_15235740 | 0.00 |
ENSRNOT00000027170
|
Mcrip2
|
MAPK regulated co-repressor interacting protein 2 |
| chr3_-_12029877 | 0.00 |
ENSRNOT00000022327
|
Slc2a8
|
solute carrier family 2 member 8 |
| chr6_+_26517840 | 0.00 |
ENSRNOT00000031842
|
Ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr20_-_3728844 | 0.00 |
ENSRNOT00000074958
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 |
| chr2_+_24024791 | 0.00 |
ENSRNOT00000089599
ENSRNOT00000066302 |
Ap3b1
|
adaptor-related protein complex 3, beta 1 subunit |
| chr11_+_34598492 | 0.00 |
ENSRNOT00000065600
|
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr12_-_39882030 | 0.00 |
ENSRNOT00000078761
ENSRNOT00000048851 |
Ppp1cc
|
protein phosphatase 1 catalytic subunit gamma |
| chr10_-_13075864 | 0.00 |
ENSRNOT00000005220
|
Paqr4
|
progestin and adipoQ receptor family member 4 |
| chr12_+_13716596 | 0.00 |
ENSRNOT00000080216
|
Actb
|
actin, beta |
| chr10_-_45798715 | 0.00 |
ENSRNOT00000028948
|
Snap47
|
synaptosomal-associated protein, 47 |
| chr7_-_144960527 | 0.00 |
ENSRNOT00000086554
|
Zfp385a
|
zinc finger protein 385A |
| chr1_+_63964155 | 0.00 |
ENSRNOT00000078512
|
AABR07002001.1
|
|
| chr1_-_229639187 | 0.00 |
ENSRNOT00000016804
|
Zfp91
|
zinc finger protein 91 |
| chr1_+_41323194 | 0.00 |
ENSRNOT00000026350
|
Esr1
|
estrogen receptor 1 |
| chr1_-_73682247 | 0.00 |
ENSRNOT00000079498
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr7_-_139254551 | 0.00 |
ENSRNOT00000083182
|
Rapgef3
|
Rap guanine nucleotide exchange factor 3 |
| chr14_-_83062302 | 0.00 |
ENSRNOT00000086769
ENSRNOT00000085735 |
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr6_-_23581052 | 0.00 |
ENSRNOT00000006190
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr10_-_89454681 | 0.00 |
ENSRNOT00000028109
|
Brca1
|
BRCA1, DNA repair associated |
| chr10_-_109827143 | 0.00 |
ENSRNOT00000054948
|
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr16_-_6404957 | 0.00 |
ENSRNOT00000048459
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr1_-_72461547 | 0.00 |
ENSRNOT00000022480
|
Ssc5d
|
scavenger receptor cysteine rich family member with 5 domains |
| chr4_-_184096806 | 0.00 |
ENSRNOT00000055433
|
LOC100362344
|
mKIAA1238 protein-like |
| chr19_+_38669230 | 0.00 |
ENSRNOT00000027273
|
Cdh3
|
cadherin 3 |
| chr10_+_12980674 | 0.00 |
ENSRNOT00000004638
|
Bicdl2
|
BICD family like cargo adaptor 2 |
| chr5_-_145418992 | 0.00 |
ENSRNOT00000037128
|
Gjb4
|
gap junction protein, beta 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) phenotypic switching(GO:0036166) |
| 0.0 | 0.1 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.0 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) positive regulation of glycogen catabolic process(GO:0045819) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |