Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
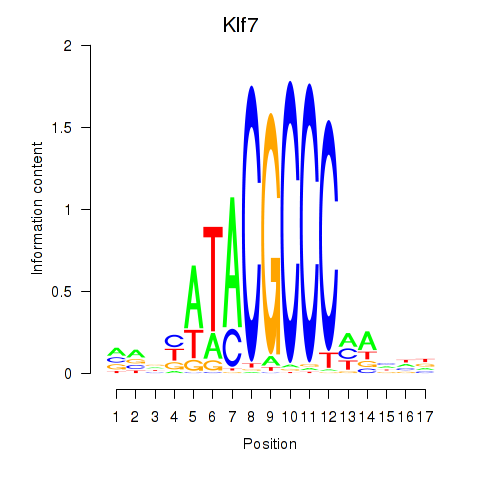
Results for Klf7
Z-value: 0.28
Transcription factors associated with Klf7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf7
|
ENSRNOG00000046242 | Kruppel like factor 7 |
|
Klf7
|
ENSRNOG00000012961 | Kruppel like factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf7 | rn6_v1_chr9_-_70787913_70787913 | 0.23 | 7.1e-01 | Click! |
Activity profile of Klf7 motif
Sorted Z-values of Klf7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_56962161 | 0.15 |
ENSRNOT00000026038
|
Alox15
|
arachidonate 15-lipoxygenase |
| chr1_-_37818879 | 0.07 |
ENSRNOT00000043747
|
LOC680200
|
similar to zinc finger protein 455 |
| chr18_+_55505993 | 0.07 |
ENSRNOT00000043736
|
RGD1309362
|
similar to interferon-inducible GTPase |
| chr10_-_38985466 | 0.06 |
ENSRNOT00000010121
|
Il13
|
interleukin 13 |
| chr3_-_165039707 | 0.06 |
ENSRNOT00000081648
|
Kcng1
|
potassium voltage-gated channel modifier subfamily G member 1 |
| chr2_+_195719543 | 0.06 |
ENSRNOT00000028324
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr1_+_162320730 | 0.06 |
ENSRNOT00000035743
|
Kctd21
|
potassium channel tetramerization domain containing 21 |
| chr12_-_17972737 | 0.05 |
ENSRNOT00000001783
|
Fam20c
|
FAM20C, golgi associated secretory pathway kinase |
| chr16_-_47207487 | 0.05 |
ENSRNOT00000059459
|
Dctd
|
dCMP deaminase |
| chr12_+_2170630 | 0.05 |
ENSRNOT00000071928
|
Pet100
|
PET100 homolog |
| chr10_+_86819929 | 0.05 |
ENSRNOT00000038228
|
Cdc6
|
cell division cycle 6 |
| chr3_-_13435979 | 0.05 |
ENSRNOT00000029600
|
Pbx3
|
PBX homeobox 3 |
| chr18_-_31136200 | 0.04 |
ENSRNOT00000084438
ENSRNOT00000080097 |
Arap3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr4_-_182844291 | 0.04 |
ENSRNOT00000064320
|
Tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr7_-_27240528 | 0.04 |
ENSRNOT00000029435
ENSRNOT00000079731 |
Hsp90b1
|
heat shock protein 90 beta family member 1 |
| chr10_+_86819472 | 0.04 |
ENSRNOT00000081424
|
Cdc6
|
cell division cycle 6 |
| chr8_-_115626380 | 0.04 |
ENSRNOT00000019107
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr9_-_98597359 | 0.04 |
ENSRNOT00000027506
|
Per2
|
period circadian clock 2 |
| chr10_-_13619935 | 0.04 |
ENSRNOT00000064392
|
Ccnf
|
cyclin F |
| chr7_+_27240876 | 0.04 |
ENSRNOT00000091725
|
LOC362863
|
first gene upstream of Nt5dc3 |
| chr17_+_45175121 | 0.04 |
ENSRNOT00000080417
|
Nkapl
|
NFKB activating protein-like |
| chr4_-_171176581 | 0.04 |
ENSRNOT00000030850
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr1_-_199655147 | 0.03 |
ENSRNOT00000026979
|
LOC103691238
|
zinc finger protein 239-like |
| chr1_+_196095214 | 0.03 |
ENSRNOT00000080741
|
LOC691716
|
similar to ribosomal protein S15a |
| chr10_+_65805860 | 0.03 |
ENSRNOT00000011795
|
Ift20
|
intraflagellar transport 20 |
| chr3_+_13838304 | 0.03 |
ENSRNOT00000025067
|
Hspa5
|
heat shock protein family A member 5 |
| chr10_+_58613674 | 0.03 |
ENSRNOT00000010975
|
Fam64a
|
family with sequence similarity 64, member A |
| chr7_-_145450233 | 0.03 |
ENSRNOT00000092974
ENSRNOT00000021523 |
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr13_+_49870976 | 0.03 |
ENSRNOT00000090170
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr8_-_115145810 | 0.03 |
ENSRNOT00000079470
|
Abhd14a
|
abhydrolase domain containing 14A |
| chr2_+_150211898 | 0.03 |
ENSRNOT00000018767
|
Sucnr1
|
succinate receptor 1 |
| chr10_-_25890639 | 0.03 |
ENSRNOT00000085499
|
Hmmr
|
hyaluronan-mediated motility receptor |
| chr7_-_134722215 | 0.03 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr3_+_150055749 | 0.03 |
ENSRNOT00000055335
|
Actl10
|
actin-like 10 |
| chr10_+_67762995 | 0.02 |
ENSRNOT00000000253
|
Zfp207
|
zinc finger protein 207 |
| chr7_-_140640953 | 0.02 |
ENSRNOT00000083156
|
Tuba1a
|
tubulin, alpha 1A |
| chr1_-_100810522 | 0.02 |
ENSRNOT00000027260
|
Atf5
|
activating transcription factor 5 |
| chr2_+_113984824 | 0.02 |
ENSRNOT00000086399
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr1_-_153740905 | 0.02 |
ENSRNOT00000023239
|
Prss23
|
protease, serine, 23 |
| chr10_-_89225297 | 0.02 |
ENSRNOT00000027868
ENSRNOT00000082010 |
Becn1
|
beclin 1 |
| chr4_-_28310178 | 0.02 |
ENSRNOT00000084021
|
RGD1563091
|
similar to OEF2 |
| chr4_-_150616895 | 0.02 |
ENSRNOT00000073562
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr20_-_8202924 | 0.02 |
ENSRNOT00000071399
|
Tmem217
|
transmembrane protein 217 |
| chr1_+_213991620 | 0.02 |
ENSRNOT00000038691
|
Rplp2
|
ribosomal protein lateral stalk subunit P2 |
| chr10_-_65805693 | 0.02 |
ENSRNOT00000012245
|
Tnfaip1
|
TNF alpha induced protein 1 |
| chr8_-_114449956 | 0.02 |
ENSRNOT00000056414
|
Col6a6
|
collagen type VI alpha 6 chain |
| chr10_-_15211325 | 0.02 |
ENSRNOT00000027083
|
Rhot2
|
ras homolog family member T2 |
| chr9_-_65329066 | 0.02 |
ENSRNOT00000018284
|
Ppil3
|
peptidylprolyl isomerase like 3 |
| chr11_-_67618704 | 0.02 |
ENSRNOT00000042208
|
Ccdc58
|
coiled-coil domain containing 58 |
| chr2_+_113984646 | 0.02 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr5_+_126254142 | 0.02 |
ENSRNOT00000057471
|
Pars2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr10_-_65437143 | 0.02 |
ENSRNOT00000017264
|
Nek8
|
NIMA-related kinase 8 |
| chr2_+_12102487 | 0.02 |
ENSRNOT00000089209
ENSRNOT00000072155 |
LOC100912538
|
centrin-3-like |
| chr10_-_88645364 | 0.02 |
ENSRNOT00000030344
|
Rab5c
|
RAB5C, member RAS oncogene family |
| chr8_+_128806129 | 0.02 |
ENSRNOT00000025225
|
Rpsa
|
ribosomal protein SA |
| chr20_+_5125679 | 0.02 |
ENSRNOT00000060832
|
Bag6
|
BCL2-associated athanogene 6 |
| chr3_-_152259156 | 0.02 |
ENSRNOT00000065729
|
LOC100910882
|
RNA-binding protein 39-like |
| chr18_+_55391388 | 0.02 |
ENSRNOT00000071612
|
LOC100910934
|
interferon-inducible GTPase 1-like |
| chr10_-_49192693 | 0.02 |
ENSRNOT00000004294
|
Zfp286a
|
zinc finger protein 286A |
| chr1_+_214428526 | 0.01 |
ENSRNOT00000057030
|
LOC100911575
|
60S acidic ribosomal protein P2-like |
| chr1_-_228263198 | 0.01 |
ENSRNOT00000028572
|
Olr1874
|
olfactory receptor 1874 |
| chr20_+_3422461 | 0.01 |
ENSRNOT00000084917
ENSRNOT00000079854 |
Tubb5
|
tubulin, beta 5 class I |
| chr8_-_77992621 | 0.01 |
ENSRNOT00000085843
|
Myzap
|
myocardial zonula adherens protein |
| chr13_-_87847263 | 0.01 |
ENSRNOT00000003650
|
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr4_-_120817377 | 0.01 |
ENSRNOT00000021826
|
Podxl2
|
podocalyxin-like 2 |
| chr12_+_22026075 | 0.01 |
ENSRNOT00000029041
|
LOC100910838
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr1_-_199716205 | 0.01 |
ENSRNOT00000027290
|
RGD1310127
|
similar to cDNA sequence BC017158 |
| chr14_-_34218961 | 0.01 |
ENSRNOT00000072588
|
LOC686911
|
similar to Exocyst complex component 1 (Exocyst complex component Sec3) |
| chr7_-_144322240 | 0.01 |
ENSRNOT00000089290
|
LOC100912282
|
calcium-binding and coiled-coil domain-containing protein 1-like |
| chr3_-_151724654 | 0.01 |
ENSRNOT00000026964
|
Rbm39
|
RNA binding motif protein 39 |
| chr18_+_59748444 | 0.01 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr10_-_25847994 | 0.01 |
ENSRNOT00000082076
|
Mat2b
|
methionine adenosyltransferase 2B |
| chr5_+_48561339 | 0.01 |
ENSRNOT00000010614
ENSRNOT00000091827 |
Rngtt
|
RNA guanylyltransferase and 5'-phosphatase |
| chr9_-_82419288 | 0.01 |
ENSRNOT00000004797
|
Tuba4a
|
tubulin, alpha 4A |
| chr7_+_12179203 | 0.01 |
ENSRNOT00000049170
|
Mbd3
|
methyl-CpG binding domain protein 3 |
| chr1_-_226630900 | 0.01 |
ENSRNOT00000028092
|
Tmem138
|
transmembrane protein 138 |
| chr6_+_86713604 | 0.01 |
ENSRNOT00000059271
|
Fam179b
|
family with sequence similarity 179, member B |
| chr5_-_25721072 | 0.01 |
ENSRNOT00000021839
|
Tmem67
|
transmembrane protein 67 |
| chr1_+_56242346 | 0.01 |
ENSRNOT00000019317
|
Smoc2
|
SPARC related modular calcium binding 2 |
| chr17_-_62462644 | 0.01 |
ENSRNOT00000024545
|
Ccny
|
cyclin Y |
| chr19_-_28296640 | 0.01 |
ENSRNOT00000041115
ENSRNOT00000042781 |
AABR07043429.1
|
|
| chr6_-_7741414 | 0.01 |
ENSRNOT00000038246
|
Thada
|
THADA, armadillo repeat containing |
| chr7_-_130176696 | 0.01 |
ENSRNOT00000051347
|
Dennd6b
|
DENN domain containing 6B |
| chr3_-_153398789 | 0.01 |
ENSRNOT00000055219
|
Mroh8
|
maestro heat-like repeat family member 8 |
| chr17_-_71897972 | 0.01 |
ENSRNOT00000065942
|
Sfmbt2
|
Scm-like with four mbt domains 2 |
| chr8_+_72029489 | 0.01 |
ENSRNOT00000089336
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chrX_+_120859968 | 0.00 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr14_-_33560503 | 0.00 |
ENSRNOT00000002862
|
Srp72
|
signal recognition particle 72 |
| chrX_+_120860178 | 0.00 |
ENSRNOT00000088661
|
Wdr44
|
WD repeat domain 44 |
| chr1_-_86948845 | 0.00 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chr12_-_38274036 | 0.00 |
ENSRNOT00000063990
|
Kntc1
|
kinetochore associated 1 |
| chr4_+_82776746 | 0.00 |
ENSRNOT00000011371
|
Tax1bp1
|
Tax1 binding protein 1 |
| chr14_+_17531398 | 0.00 |
ENSRNOT00000050038
|
Cdkl2
|
cyclin dependent kinase like 2 |
| chr10_-_15161938 | 0.00 |
ENSRNOT00000026641
|
Fam173a
|
family with sequence similarity 173, member A |
| chr13_-_78521911 | 0.00 |
ENSRNOT00000087506
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr8_+_128577080 | 0.00 |
ENSRNOT00000024172
|
Wdr48
|
WD repeat domain 48 |
| chr3_-_79743737 | 0.00 |
ENSRNOT00000013584
|
Ptpmt1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr6_+_86713803 | 0.00 |
ENSRNOT00000005861
|
Fam179b
|
family with sequence similarity 179, member B |
| chr20_-_2040422 | 0.00 |
ENSRNOT00000052311
|
RT1-M5
|
RT1 class Ib, locus M5 |
| chr4_+_7835949 | 0.00 |
ENSRNOT00000065962
|
Tomm7
|
translocase of outer mitochondrial membrane 7 |
| chr10_+_25890943 | 0.00 |
ENSRNOT00000086814
|
Nudcd2
|
NudC domain containing 2 |
| chrX_+_6430594 | 0.00 |
ENSRNOT00000044009
|
Maob
|
monoamine oxidase B |
| chr1_-_167308827 | 0.00 |
ENSRNOT00000027590
ENSRNOT00000027575 |
Nup98
|
nucleoporin 98 |
| chr19_-_29802083 | 0.00 |
ENSRNOT00000024755
|
AC123471.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) dentinogenesis(GO:0097187) |
| 0.0 | 0.0 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.1 | GO:1904117 | cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.0 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |