Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Klf15
Z-value: 0.84
Transcription factors associated with Klf15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf15
|
ENSRNOG00000017808 | Kruppel-like factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf15 | rn6_v1_chr4_+_122365093_122365093 | 0.06 | 9.3e-01 | Click! |
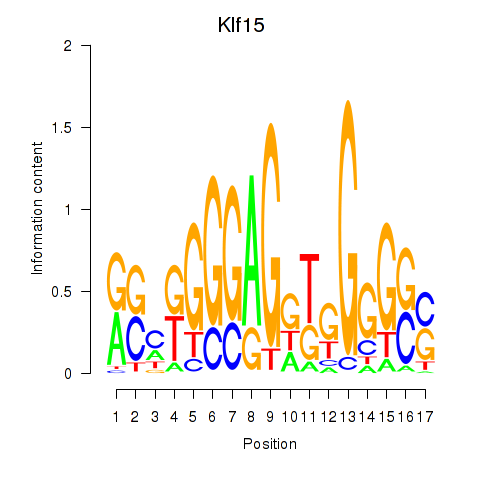
Activity profile of Klf15 motif
Sorted Z-values of Klf15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_226435979 | 0.67 |
ENSRNOT00000048704
ENSRNOT00000036232 ENSRNOT00000035576 ENSRNOT00000036180 ENSRNOT00000036168 ENSRNOT00000047964 ENSRNOT00000036283 ENSRNOT00000007429 |
Syt7
|
synaptotagmin 7 |
| chr10_-_85517683 | 0.48 |
ENSRNOT00000016070
|
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr12_-_16002788 | 0.45 |
ENSRNOT00000090318
|
Amz1
|
archaelysin family metallopeptidase 1 |
| chr20_-_5064469 | 0.37 |
ENSRNOT00000001120
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr7_+_130474279 | 0.35 |
ENSRNOT00000092388
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr3_+_41019898 | 0.32 |
ENSRNOT00000007335
|
Kcnj3
|
potassium voltage-gated channel subfamily J member 3 |
| chr5_-_172623899 | 0.28 |
ENSRNOT00000080591
|
Ski
|
SKI proto-oncogene |
| chr15_+_34520142 | 0.28 |
ENSRNOT00000074659
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr12_-_36398206 | 0.27 |
ENSRNOT00000090944
ENSRNOT00000058492 |
Tmem132b
|
transmembrane protein 132B |
| chr18_-_86878142 | 0.26 |
ENSRNOT00000058139
|
Dok6
|
docking protein 6 |
| chr15_+_120372 | 0.23 |
ENSRNOT00000007828
|
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr3_-_7422738 | 0.23 |
ENSRNOT00000088339
|
Gtf3c4
|
general transcription factor IIIC subunit 4 |
| chr19_-_41798383 | 0.22 |
ENSRNOT00000021744
|
Phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr3_-_1924827 | 0.22 |
ENSRNOT00000006162
|
Cacna1b
|
calcium voltage-gated channel subunit alpha1 B |
| chrX_+_71155601 | 0.22 |
ENSRNOT00000076453
ENSRNOT00000048521 |
Foxo4
|
forkhead box O4 |
| chr3_+_58632476 | 0.21 |
ENSRNOT00000010630
|
Rapgef4
|
Rap guanine nucleotide exchange factor 4 |
| chr3_-_164095878 | 0.21 |
ENSRNOT00000079414
|
B4galt5
|
beta-1,4-galactosyltransferase 5 |
| chrX_-_22440187 | 0.21 |
ENSRNOT00000090731
|
Gpr173
|
|
| chr20_+_4363508 | 0.19 |
ENSRNOT00000077205
|
Ager
|
advanced glycosylation end product-specific receptor |
| chr4_-_115157263 | 0.19 |
ENSRNOT00000015296
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr5_+_43603043 | 0.18 |
ENSRNOT00000009899
|
Epha7
|
Eph receptor A7 |
| chr7_+_130474508 | 0.18 |
ENSRNOT00000085191
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr16_+_20740826 | 0.18 |
ENSRNOT00000038057
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chr20_+_5646097 | 0.17 |
ENSRNOT00000090925
|
Itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr5_-_147412705 | 0.17 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr7_-_139318455 | 0.17 |
ENSRNOT00000092029
|
Hdac7
|
histone deacetylase 7 |
| chr5_+_139783951 | 0.17 |
ENSRNOT00000081333
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr10_+_66690133 | 0.16 |
ENSRNOT00000046262
|
Nf1
|
neurofibromin 1 |
| chr10_+_59360765 | 0.16 |
ENSRNOT00000036278
|
Zzef1
|
zinc finger ZZ-type and EF-hand domain containing 1 |
| chr2_+_230901126 | 0.16 |
ENSRNOT00000016026
ENSRNOT00000015564 ENSRNOT00000068198 |
Camk2d
|
calcium/calmodulin-dependent protein kinase II delta |
| chr5_-_138545404 | 0.16 |
ENSRNOT00000011586
|
Rimkla
|
ribosomal modification protein rimK-like family member A |
| chr5_-_100647727 | 0.16 |
ENSRNOT00000067435
|
Nfib
|
nuclear factor I/B |
| chr5_+_74649765 | 0.15 |
ENSRNOT00000075952
|
Palm2
|
paralemmin 2 |
| chr13_-_52256196 | 0.15 |
ENSRNOT00000011105
|
Ipo9
|
importin 9 |
| chr7_-_143863186 | 0.15 |
ENSRNOT00000017096
|
Rarg
|
retinoic acid receptor, gamma |
| chr4_+_133286114 | 0.15 |
ENSRNOT00000084158
|
Ppp4r2
|
protein phosphatase 4, regulatory subunit 2 |
| chrX_+_70596901 | 0.15 |
ENSRNOT00000088114
|
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr1_+_80000165 | 0.15 |
ENSRNOT00000084912
|
Six5
|
SIX homeobox 5 |
| chr7_+_97559841 | 0.15 |
ENSRNOT00000007326
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr2_+_198823366 | 0.14 |
ENSRNOT00000083769
ENSRNOT00000028814 |
Pias3
|
protein inhibitor of activated STAT, 3 |
| chr13_+_34365147 | 0.14 |
ENSRNOT00000093066
|
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr14_+_83510278 | 0.14 |
ENSRNOT00000081161
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr1_-_164590562 | 0.13 |
ENSRNOT00000024157
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr16_+_60925093 | 0.13 |
ENSRNOT00000015813
|
Tnks
|
tankyrase |
| chr11_-_38457373 | 0.13 |
ENSRNOT00000041177
|
Zfp295
|
zinc finger protein 295 |
| chr19_+_25181564 | 0.13 |
ENSRNOT00000008104
|
Rfx1
|
regulatory factor X1 |
| chr11_-_83926524 | 0.12 |
ENSRNOT00000041777
ENSRNOT00000040029 |
Eif4g1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr3_-_80003032 | 0.12 |
ENSRNOT00000017360
|
Madd
|
MAP-kinase activating death domain |
| chr3_-_57119001 | 0.12 |
ENSRNOT00000083171
|
Tlk1
|
tousled-like kinase 1 |
| chr9_+_88357556 | 0.11 |
ENSRNOT00000020669
|
Col4a3
|
collagen type IV alpha 3 chain |
| chr9_-_16979044 | 0.11 |
ENSRNOT00000073191
ENSRNOT00000025335 |
Zfp318
|
zinc finger protein 318 |
| chr8_+_79159370 | 0.11 |
ENSRNOT00000086841
|
Rfx7
|
regulatory factor X, 7 |
| chr14_+_60857989 | 0.11 |
ENSRNOT00000034411
|
Ccdc149
|
coiled-coil domain containing 149 |
| chr11_+_24263281 | 0.10 |
ENSRNOT00000086946
|
Gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr5_+_63056089 | 0.10 |
ENSRNOT00000081090
|
Tgfbr1
|
transforming growth factor, beta receptor 1 |
| chr14_-_82287706 | 0.10 |
ENSRNOT00000080695
|
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr20_+_4363152 | 0.10 |
ENSRNOT00000000508
ENSRNOT00000084841 ENSRNOT00000072848 ENSRNOT00000077561 |
Ager
|
advanced glycosylation end product-specific receptor |
| chr6_-_50943488 | 0.10 |
ENSRNOT00000068419
|
Dus4l
|
dihydrouridine synthase 4-like |
| chr7_+_122818975 | 0.10 |
ENSRNOT00000000206
|
Ep300
|
E1A binding protein p300 |
| chr6_-_111329882 | 0.09 |
ENSRNOT00000074480
|
Ism2
|
isthmin 2 |
| chr8_+_117737387 | 0.09 |
ENSRNOT00000090164
ENSRNOT00000091573 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr7_+_123381077 | 0.09 |
ENSRNOT00000082603
ENSRNOT00000056041 |
Srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr10_+_90550147 | 0.09 |
ENSRNOT00000032944
|
Fzd2
|
frizzled class receptor 2 |
| chr8_+_56179637 | 0.08 |
ENSRNOT00000035989
|
Arhgap20
|
Rho GTPase activating protein 20 |
| chr1_+_78735678 | 0.08 |
ENSRNOT00000089715
|
Strn4
|
striatin 4 |
| chr17_-_85141210 | 0.08 |
ENSRNOT00000000162
|
Dnajc1
|
DnaJ heat shock protein family (Hsp40) member C1 |
| chr17_-_10640548 | 0.08 |
ENSRNOT00000064274
|
Simc1
|
SUMO-interacting motifs containing 1 |
| chr7_-_90318221 | 0.08 |
ENSRNOT00000050774
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr1_-_219532609 | 0.08 |
ENSRNOT00000025641
|
Ankrd13d
|
ankyrin repeat domain 13D |
| chr8_-_55408464 | 0.08 |
ENSRNOT00000066893
|
Sik2
|
salt-inducible kinase 2 |
| chr7_-_77162148 | 0.07 |
ENSRNOT00000008350
|
Klf10
|
Kruppel-like factor 10 |
| chr1_-_57327379 | 0.07 |
ENSRNOT00000080429
|
Dll1
|
delta like canonical Notch ligand 1 |
| chr8_+_117780891 | 0.07 |
ENSRNOT00000077236
|
Shisa5
|
shisa family member 5 |
| chr15_+_34493138 | 0.07 |
ENSRNOT00000089584
ENSRNOT00000027789 |
Nfatc4
|
nuclear factor of activated T-cells 4 |
| chr8_-_116965396 | 0.07 |
ENSRNOT00000042528
|
Bsn
|
bassoon (presynaptic cytomatrix protein) |
| chr8_-_64268555 | 0.07 |
ENSRNOT00000013581
ENSRNOT00000084758 |
Arih1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr3_-_91839009 | 0.07 |
ENSRNOT00000083703
|
Ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr7_-_70969905 | 0.06 |
ENSRNOT00000057745
|
Nab2
|
Ngfi-A binding protein 2 |
| chr14_-_84106997 | 0.06 |
ENSRNOT00000065501
|
Osbp2
|
oxysterol binding protein 2 |
| chr3_+_7422820 | 0.06 |
ENSRNOT00000064323
|
Ddx31
|
DEAD-box helicase 31 |
| chr10_-_82197848 | 0.06 |
ENSRNOT00000081307
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr10_-_64862268 | 0.06 |
ENSRNOT00000056234
|
Phf12
|
PHD finger protein 12 |
| chr18_-_47513030 | 0.06 |
ENSRNOT00000083881
ENSRNOT00000074226 |
Lox
|
lysyl oxidase |
| chr4_-_66002444 | 0.06 |
ENSRNOT00000018645
|
Zc3hav1l
|
zinc finger CCCH-type containing, antiviral 1 like |
| chr10_+_59585072 | 0.06 |
ENSRNOT00000025009
|
Camkk1
|
calcium/calmodulin-dependent protein kinase kinase 1 |
| chr19_+_38039564 | 0.06 |
ENSRNOT00000087491
|
Nfatc3
|
nuclear factor of activated T-cells 3 |
| chr4_+_153217782 | 0.06 |
ENSRNOT00000015499
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr13_-_74077783 | 0.06 |
ENSRNOT00000005677
|
Soat1
|
sterol O-acyltransferase 1 |
| chr13_+_89524329 | 0.06 |
ENSRNOT00000004279
|
Mpz
|
myelin protein zero |
| chr1_+_199225100 | 0.05 |
ENSRNOT00000088606
|
Setd1a
|
SET domain containing 1A |
| chr6_+_28235695 | 0.05 |
ENSRNOT00000047210
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr5_-_100647298 | 0.05 |
ENSRNOT00000067538
ENSRNOT00000013092 |
Nfib
|
nuclear factor I/B |
| chr17_-_80320681 | 0.05 |
ENSRNOT00000023637
|
C1ql3
|
complement C1q like 3 |
| chr2_+_207108552 | 0.05 |
ENSRNOT00000027234
|
Slc16a1
|
solute carrier family 16 member 1 |
| chr8_+_117068582 | 0.05 |
ENSRNOT00000073559
|
Amt
|
aminomethyltransferase |
| chr2_+_189615948 | 0.04 |
ENSRNOT00000092144
|
Crtc2
|
CREB regulated transcription coactivator 2 |
| chr12_-_22417980 | 0.04 |
ENSRNOT00000072208
|
Ephb4
|
EPH receptor B4 |
| chr1_+_282265370 | 0.04 |
ENSRNOT00000015687
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr5_-_82168347 | 0.04 |
ENSRNOT00000084959
ENSRNOT00000084147 |
Astn2
|
astrotactin 2 |
| chr1_+_78800754 | 0.04 |
ENSRNOT00000084601
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr6_-_115352681 | 0.04 |
ENSRNOT00000005873
|
Gtf2a1
|
general transcription factor 2A subunit 1 |
| chr6_-_46631983 | 0.04 |
ENSRNOT00000045963
|
Sox11
|
SRY box 11 |
| chr2_-_29121104 | 0.04 |
ENSRNOT00000020543
|
Tnpo1
|
transportin 1 |
| chr6_+_33786197 | 0.03 |
ENSRNOT00000082598
|
Pum2
|
pumilio RNA-binding family member 2 |
| chr10_-_108691367 | 0.03 |
ENSRNOT00000005067
|
Nptx1
|
neuronal pentraxin 1 |
| chr9_+_93326283 | 0.03 |
ENSRNOT00000024578
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr4_-_83137527 | 0.03 |
ENSRNOT00000039580
|
Jazf1
|
JAZF zinc finger 1 |
| chr1_+_23977688 | 0.03 |
ENSRNOT00000014805
|
Tbpl1
|
TATA-box binding protein like 1 |
| chr8_+_117737117 | 0.03 |
ENSRNOT00000028039
|
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr10_-_88667345 | 0.03 |
ENSRNOT00000025518
|
Kcnh4
|
potassium voltage-gated channel subfamily H member 4 |
| chr11_-_61499557 | 0.03 |
ENSRNOT00000046208
|
Usf3
|
upstream transcription factor family member 3 |
| chr5_-_169167831 | 0.03 |
ENSRNOT00000012407
|
Phf13
|
PHD finger protein 13 |
| chr4_-_152380023 | 0.03 |
ENSRNOT00000012397
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr1_+_174702373 | 0.02 |
ENSRNOT00000013733
|
Zfp143
|
zinc finger protein 143 |
| chrX_-_123254557 | 0.02 |
ENSRNOT00000039809
|
Akap17b
|
A kinase (PRKA) anchor protein 17B |
| chr16_-_6404578 | 0.02 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr20_-_32052855 | 0.02 |
ENSRNOT00000074989
|
LOC100364027
|
hexokinase 1-like |
| chr8_+_97277456 | 0.02 |
ENSRNOT00000082757
|
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr19_+_38039729 | 0.02 |
ENSRNOT00000089783
|
Nfatc3
|
nuclear factor of activated T-cells 3 |
| chr2_+_62150493 | 0.02 |
ENSRNOT00000080241
|
Zfr
|
zinc finger RNA binding protein |
| chr8_+_130581062 | 0.02 |
ENSRNOT00000014323
|
Fam198a
|
family with sequence similarity 198, member A |
| chr1_-_199395363 | 0.02 |
ENSRNOT00000090368
ENSRNOT00000026587 |
Prss36
|
protease, serine, 36 |
| chr1_+_101610773 | 0.02 |
ENSRNOT00000032535
|
Rasip1
|
Ras interacting protein 1 |
| chr10_-_59360661 | 0.02 |
ENSRNOT00000036942
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr7_-_143966863 | 0.02 |
ENSRNOT00000018828
|
Sp7
|
Sp7 transcription factor |
| chr17_-_9519595 | 0.02 |
ENSRNOT00000029885
|
Ddx46
|
DEAD-box helicase 46 |
| chr10_-_85124644 | 0.02 |
ENSRNOT00000012376
|
Kpnb1
|
karyopherin subunit beta 1 |
| chrX_-_118615798 | 0.01 |
ENSRNOT00000045463
|
Lrch2
|
leucine rich repeats and calponin homology domain containing 2 |
| chr5_+_166533181 | 0.01 |
ENSRNOT00000045063
|
Clstn1
|
calsyntenin 1 |
| chr8_+_45797315 | 0.01 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chr4_-_152380184 | 0.01 |
ENSRNOT00000091473
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr1_+_221099998 | 0.01 |
ENSRNOT00000028262
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chrX_+_112769645 | 0.01 |
ENSRNOT00000064478
|
Col4a5
|
collagen type IV alpha 5 chain |
| chr18_-_325374 | 0.01 |
ENSRNOT00000078032
ENSRNOT00000003638 |
Mtmr1
|
myotubularin related protein 1 |
| chr4_+_133285552 | 0.01 |
ENSRNOT00000029115
|
Ppp4r2
|
protein phosphatase 4, regulatory subunit 2 |
| chrX_+_27015884 | 0.01 |
ENSRNOT00000065814
|
Msl3
|
male-specific lethal 3 homolog (Drosophila) |
| chr6_-_111295917 | 0.01 |
ENSRNOT00000073428
ENSRNOT00000079373 |
Vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr1_+_140762758 | 0.01 |
ENSRNOT00000047848
|
Acan
|
aggrecan |
| chr18_+_28781349 | 0.01 |
ENSRNOT00000026112
|
Psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr1_-_220786615 | 0.00 |
ENSRNOT00000038198
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr1_+_226091774 | 0.00 |
ENSRNOT00000027693
|
Fads3
|
fatty acid desaturase 3 |
| chr9_-_92435363 | 0.00 |
ENSRNOT00000093735
ENSRNOT00000022822 ENSRNOT00000093245 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr4_-_71713063 | 0.00 |
ENSRNOT00000059447
|
Fam131b
|
family with sequence similarity 131, member B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf15
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.5 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.1 | 0.3 | GO:1905204 | response to selenite ion(GO:0072714) negative regulation of connective tissue replacement(GO:1905204) |
| 0.1 | 0.2 | GO:0021896 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.1 | 0.2 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.2 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.1 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.3 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 0.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.2 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.1 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0036395 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) |
| 0.0 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:1905075 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.3 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.1 | GO:0018076 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.2 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0045608 | inhibition of neuroepithelial cell differentiation(GO:0002085) cerebellar cortex structural organization(GO:0021698) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.5 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.3 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.1 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 | 0.0 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0060170 | ciliary membrane(GO:0060170) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.3 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.1 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.2 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |