Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
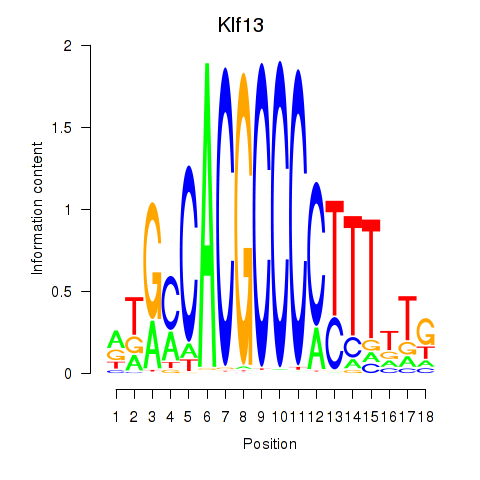
Results for Klf13
Z-value: 0.31
Transcription factors associated with Klf13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf13
|
ENSRNOG00000015822 | Kruppel-like factor 13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf13 | rn6_v1_chr1_-_124803363_124803363 | 0.80 | 1.0e-01 | Click! |
Activity profile of Klf13 motif
Sorted Z-values of Klf13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_14395362 | 0.31 |
ENSRNOT00000092787
|
Rab14
|
RAB14, member RAS oncogene family |
| chr1_-_78212350 | 0.17 |
ENSRNOT00000071098
|
Inafm1
|
InaF-motif containing 1 |
| chr19_+_10563423 | 0.12 |
ENSRNOT00000021037
|
Dok4
|
docking protein 4 |
| chr2_+_122877286 | 0.12 |
ENSRNOT00000033080
|
Arse
|
arylsulfatase E |
| chr16_-_19894591 | 0.11 |
ENSRNOT00000085940
|
Ano8
|
anoctamin 8 |
| chr19_+_57047830 | 0.10 |
ENSRNOT00000080860
|
Galnt2
|
polypeptide N-acetylgalactosaminyltransferase 2 |
| chr20_+_3351303 | 0.09 |
ENSRNOT00000080419
ENSRNOT00000001065 ENSRNOT00000086503 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr20_+_5049496 | 0.09 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr12_-_24046814 | 0.08 |
ENSRNOT00000001961
|
Por
|
cytochrome p450 oxidoreductase |
| chr19_+_26106838 | 0.07 |
ENSRNOT00000035987
|
Hook2
|
hook microtubule-tethering protein 2 |
| chr7_+_11414446 | 0.06 |
ENSRNOT00000027441
|
Pias4
|
protein inhibitor of activated STAT, 4 |
| chr1_+_219345918 | 0.05 |
ENSRNOT00000025018
|
Cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr4_+_171748273 | 0.05 |
ENSRNOT00000009998
|
Dera
|
deoxyribose-phosphate aldolase |
| chr7_+_12619774 | 0.04 |
ENSRNOT00000015257
|
Med16
|
mediator complex subunit 16 |
| chr5_-_58484900 | 0.04 |
ENSRNOT00000012386
|
Fam214b
|
family with sequence similarity 214, member B |
| chr12_-_19582185 | 0.04 |
ENSRNOT00000060020
ENSRNOT00000085853 |
RGD1305455
|
similar to hypothetical protein FLJ10925 |
| chr4_-_117153907 | 0.04 |
ENSRNOT00000091374
|
Rab11fip5
|
RAB11 family interacting protein 5 |
| chr5_-_150506871 | 0.04 |
ENSRNOT00000086131
|
Trnau1ap
|
tRNA selenocysteine 1 associated protein 1 |
| chr17_+_15845931 | 0.04 |
ENSRNOT00000092083
|
Card19
|
caspase recruitment domain family, member 19 |
| chr12_+_22026075 | 0.03 |
ENSRNOT00000029041
|
LOC100910838
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr12_-_19599374 | 0.03 |
ENSRNOT00000001849
|
Gpc2
|
glypican 2 |
| chr20_-_10912769 | 0.03 |
ENSRNOT00000051678
|
RGD1566085
|
similar to pyridoxal (pyridoxine, vitamin B6) kinase |
| chr12_+_47218969 | 0.03 |
ENSRNOT00000081343
ENSRNOT00000038395 |
Unc119b
|
unc-119 lipid binding chaperone B |
| chr14_+_44479614 | 0.03 |
ENSRNOT00000003691
|
Ugdh
|
UDP-glucose 6-dehydrogenase |
| chr2_+_251453644 | 0.03 |
ENSRNOT00000041897
|
Znhit6
|
zinc finger, HIT-type containing 6 |
| chr13_-_79801561 | 0.03 |
ENSRNOT00000075936
|
Suco
|
SUN domain containing ossification factor |
| chr3_+_164274710 | 0.03 |
ENSRNOT00000012939
|
Snai1
|
snail family transcriptional repressor 1 |
| chr20_-_5533448 | 0.03 |
ENSRNOT00000000568
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr20_-_5533600 | 0.03 |
ENSRNOT00000072319
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr6_-_39363367 | 0.02 |
ENSRNOT00000088687
ENSRNOT00000065531 |
Fam84a
|
family with sequence similarity 84, member A |
| chr4_-_116786391 | 0.02 |
ENSRNOT00000086297
ENSRNOT00000091490 |
Exoc6b
|
exocyst complex component 6B |
| chr1_+_166739532 | 0.02 |
ENSRNOT00000079846
ENSRNOT00000026665 |
Clpb
|
ClpB homolog, mitochondrial AAA ATPase chaperonin |
| chr8_+_64364741 | 0.02 |
ENSRNOT00000082840
|
Celf6
|
CUGBP, Elav-like family member 6 |
| chr3_-_176144531 | 0.02 |
ENSRNOT00000082266
|
Tcfl5
|
transcription factor like 5 |
| chr20_-_5155293 | 0.02 |
ENSRNOT00000092322
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr10_+_90230441 | 0.02 |
ENSRNOT00000082722
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr7_-_122963299 | 0.02 |
ENSRNOT00000090208
|
Rangap1
|
RAN GTPase activating protein 1 |
| chr19_+_24747178 | 0.02 |
ENSRNOT00000038879
ENSRNOT00000079106 |
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr12_+_2069959 | 0.02 |
ENSRNOT00000001298
|
Pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr12_+_2070365 | 0.02 |
ENSRNOT00000072952
|
Pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr3_+_164275095 | 0.02 |
ENSRNOT00000077581
|
Snai1
|
snail family transcriptional repressor 1 |
| chr12_-_44279002 | 0.02 |
ENSRNOT00000064900
|
Fbxo21
|
F-box protein 21 |
| chr5_+_60572585 | 0.02 |
ENSRNOT00000029704
|
Polr1e
|
RNA polymerase I subunit E |
| chr3_-_60795951 | 0.02 |
ENSRNOT00000002174
|
Atf2
|
activating transcription factor 2 |
| chr4_-_51726212 | 0.02 |
ENSRNOT00000009261
|
Wasl
|
Wiskott-Aldrich syndrome-like |
| chr7_+_31784438 | 0.01 |
ENSRNOT00000010914
ENSRNOT00000010929 |
Ikbip
|
IKBKB interacting protein |
| chr1_-_101500850 | 0.01 |
ENSRNOT00000028390
|
Nucb1
|
nucleobindin 1 |
| chr5_-_159583049 | 0.01 |
ENSRNOT00000055839
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr6_-_102353403 | 0.01 |
ENSRNOT00000090407
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr4_+_7662019 | 0.01 |
ENSRNOT00000014023
|
Fam126a
|
family with sequence similarity 126, member A |
| chr1_+_80141630 | 0.01 |
ENSRNOT00000029552
|
Opa3
|
optic atrophy 3 |
| chr20_-_49486550 | 0.01 |
ENSRNOT00000048270
ENSRNOT00000076541 |
Prdm1
|
PR/SET domain 1 |
| chr9_-_43022998 | 0.01 |
ENSRNOT00000063781
ENSRNOT00000089843 |
Lman2l
|
lectin, mannose-binding 2-like |
| chr9_+_113699170 | 0.01 |
ENSRNOT00000017915
|
Twsg1
|
twisted gastrulation BMP signaling modulator 1 |
| chr18_-_31094303 | 0.01 |
ENSRNOT00000078667
|
Hdac3
|
histone deacetylase 3 |
| chr5_+_154489590 | 0.01 |
ENSRNOT00000035788
|
Id3
|
inhibitor of DNA binding 3, HLH protein |
| chr5_+_159428515 | 0.01 |
ENSRNOT00000010183
|
Padi2
|
peptidyl arginine deiminase 2 |
| chr5_+_64476317 | 0.01 |
ENSRNOT00000017217
|
LOC108348074
|
collagen alpha-1(XV) chain-like |
| chr2_-_112831476 | 0.01 |
ENSRNOT00000018055
|
Ect2
|
epithelial cell transforming 2 |
| chr12_+_25036605 | 0.01 |
ENSRNOT00000001996
ENSRNOT00000084427 |
Limk1
|
LIM domain kinase 1 |
| chr13_-_79801112 | 0.01 |
ENSRNOT00000087323
ENSRNOT00000036483 |
Suco
|
SUN domain containing ossification factor |
| chr13_+_44957014 | 0.01 |
ENSRNOT00000004878
|
Ubxn4
|
UBX domain protein 4 |
| chr20_+_10265806 | 0.01 |
ENSRNOT00000001564
ENSRNOT00000086272 |
Ndufv3
|
NADH:ubiquinone oxidoreductase subunit V3 |
| chr7_-_31784192 | 0.01 |
ENSRNOT00000010869
|
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr12_-_39641285 | 0.00 |
ENSRNOT00000001729
|
Anapc7
|
anaphase promoting complex subunit 7 |
| chr5_+_138470069 | 0.00 |
ENSRNOT00000076343
ENSRNOT00000064073 |
Zmynd12
|
zinc finger, MYND-type containing 12 |
| chr9_-_81879211 | 0.00 |
ENSRNOT00000085140
|
Rnf25
|
ring finger protein 25 |
| chr4_-_113497396 | 0.00 |
ENSRNOT00000079173
ENSRNOT00000067314 |
Pole4
|
DNA polymerase epsilon 4, accessory subunit |
| chr2_-_260148589 | 0.00 |
ENSRNOT00000013238
|
Acadm
|
acyl-CoA dehydrogenase, C-4 to C-12 straight chain |
| chr11_+_42858478 | 0.00 |
ENSRNOT00000002293
|
Arl6
|
ADP-ribosylation factor like GTPase 6 |
| chr4_+_115473811 | 0.00 |
ENSRNOT00000042875
|
Nagk
|
N-acetylglucosamine kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0042126 | growth plate cartilage chondrocyte proliferation(GO:0003419) nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |