Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
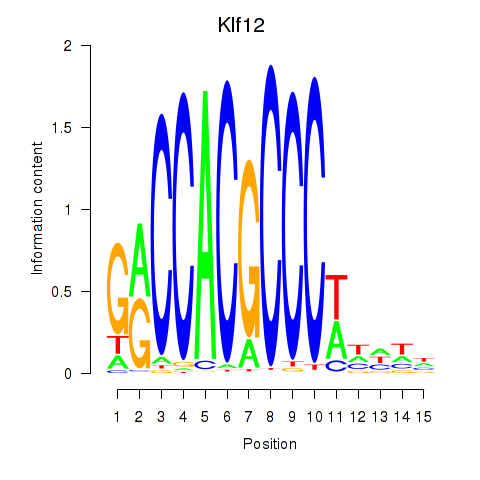
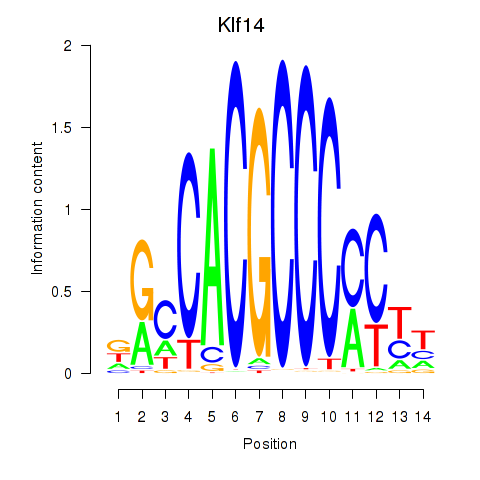
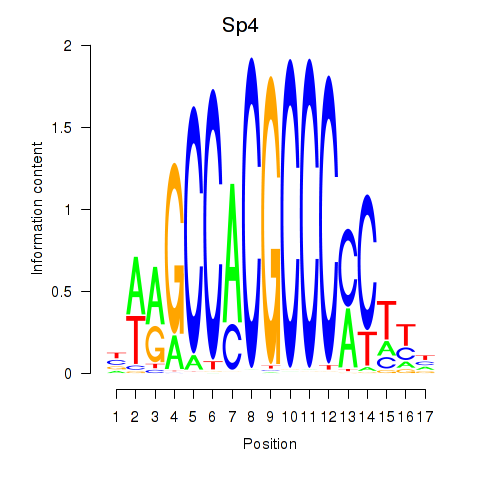
Results for Klf12_Klf14_Sp4
Z-value: 0.71
Transcription factors associated with Klf12_Klf14_Sp4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf12
|
ENSRNOG00000009145 | Kruppel-like factor 12 |
|
Klf14
|
ENSRNOG00000027557 | Kruppel-like factor 14 |
|
Sp4
|
ENSRNOG00000005472 | Sp4 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf12 | rn6_v1_chr15_-_84748525_84748525 | -0.77 | 1.3e-01 | Click! |
| Klf14 | rn6_v1_chr4_-_58250798_58250798 | 0.74 | 1.5e-01 | Click! |
| Sp4 | rn6_v1_chr6_-_146195819_146195819 | -0.15 | 8.1e-01 | Click! |
Activity profile of Klf12_Klf14_Sp4 motif
Sorted Z-values of Klf12_Klf14_Sp4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_78212350 | 1.50 |
ENSRNOT00000071098
|
Inafm1
|
InaF-motif containing 1 |
| chr10_-_56962161 | 0.89 |
ENSRNOT00000026038
|
Alox15
|
arachidonate 15-lipoxygenase |
| chr3_-_14395362 | 0.62 |
ENSRNOT00000092787
|
Rab14
|
RAB14, member RAS oncogene family |
| chr20_-_29511382 | 0.57 |
ENSRNOT00000085026
|
Ddit4
|
DNA-damage-inducible transcript 4 |
| chr14_-_82055290 | 0.56 |
ENSRNOT00000058062
|
RGD1560394
|
RGD1560394 |
| chr3_-_9037942 | 0.55 |
ENSRNOT00000036770
|
Ier5l
|
immediate early response 5-like |
| chr12_-_16934706 | 0.46 |
ENSRNOT00000001720
|
Mafk
|
MAF bZIP transcription factor K |
| chr20_-_56197025 | 0.44 |
ENSRNOT00000073028
|
Cd99
|
CD99 molecule |
| chrX_-_155862363 | 0.39 |
ENSRNOT00000089847
|
Dkc1
|
dyskerin pseudouridine synthase 1 |
| chr20_+_12429315 | 0.37 |
ENSRNOT00000001675
|
Pcbp3
|
poly(rC) binding protein 3 |
| chr16_+_20352480 | 0.36 |
ENSRNOT00000025956
|
Arrdc2
|
arrestin domain containing 2 |
| chr12_+_12227010 | 0.35 |
ENSRNOT00000060843
ENSRNOT00000092610 |
Baiap2l1
|
BAI1-associated protein 2-like 1 |
| chr1_+_80279706 | 0.33 |
ENSRNOT00000047105
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr20_+_5049496 | 0.32 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr2_-_25235275 | 0.32 |
ENSRNOT00000061580
|
F2rl1
|
F2R like trypsin receptor 1 |
| chr7_+_120580743 | 0.31 |
ENSRNOT00000017181
|
Maff
|
MAF bZIP transcription factor F |
| chr12_-_30304036 | 0.30 |
ENSRNOT00000001218
|
Nupr2
|
nuclear protein 2, transcriptional regulator |
| chr4_-_123510217 | 0.29 |
ENSRNOT00000080734
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr5_+_151413382 | 0.29 |
ENSRNOT00000012626
|
Cd164l2
|
CD164 molecule like 2 |
| chr5_-_153625869 | 0.28 |
ENSRNOT00000024464
|
Clic4
|
chloride intracellular channel 4 |
| chr9_-_9702306 | 0.27 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr16_-_20860767 | 0.26 |
ENSRNOT00000027275
ENSRNOT00000092417 |
Gdf1
Cers1
|
growth differentiation factor 1 ceramide synthase 1 |
| chr12_+_22641104 | 0.26 |
ENSRNOT00000001916
|
Serpine1
|
serpin family E member 1 |
| chr8_+_64364741 | 0.26 |
ENSRNOT00000082840
|
Celf6
|
CUGBP, Elav-like family member 6 |
| chr20_+_6351458 | 0.26 |
ENSRNOT00000091731
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr3_-_14538241 | 0.26 |
ENSRNOT00000025904
|
Stom
|
stomatin |
| chr7_-_11648322 | 0.26 |
ENSRNOT00000026871
|
Gadd45b
|
growth arrest and DNA-damage-inducible, beta |
| chr1_-_107373807 | 0.25 |
ENSRNOT00000056024
|
Svip
|
small VCP interacting protein |
| chr10_-_15166457 | 0.25 |
ENSRNOT00000026676
|
Metrn
|
meteorin, glial cell differentiation regulator |
| chr15_+_40937708 | 0.23 |
ENSRNOT00000018407
|
Spata13
|
spermatogenesis associated 13 |
| chr2_+_122877286 | 0.23 |
ENSRNOT00000033080
|
Arse
|
arylsulfatase E |
| chr1_+_220826560 | 0.22 |
ENSRNOT00000027891
|
Fosl1
|
FOS like 1, AP-1 transcription factor subunit |
| chr1_-_47213749 | 0.22 |
ENSRNOT00000024656
|
Dynlt1
|
dynein light chain Tctex-type 1 |
| chr7_+_11414446 | 0.22 |
ENSRNOT00000027441
|
Pias4
|
protein inhibitor of activated STAT, 4 |
| chr10_+_55707164 | 0.22 |
ENSRNOT00000009757
|
Hes7
|
hes family bHLH transcription factor 7 |
| chr18_-_28454756 | 0.21 |
ENSRNOT00000040091
|
Spata24
|
spermatogenesis associated 24 |
| chr20_+_6102277 | 0.21 |
ENSRNOT00000033064
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr14_-_83776863 | 0.21 |
ENSRNOT00000026481
|
Smtn
|
smoothelin |
| chr12_-_24046814 | 0.21 |
ENSRNOT00000001961
|
Por
|
cytochrome p450 oxidoreductase |
| chr7_-_18554227 | 0.21 |
ENSRNOT00000036172
ENSRNOT00000011047 |
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr10_-_38782419 | 0.21 |
ENSRNOT00000073964
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr11_+_87204175 | 0.20 |
ENSRNOT00000000306
|
Slc25a1
|
solute carrier family 25 member 1 |
| chr1_+_163445527 | 0.20 |
ENSRNOT00000020520
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr11_-_36479868 | 0.20 |
ENSRNOT00000075762
|
LOC100911295
|
non-histone chromosomal protein HMG-14-like |
| chr7_+_12974169 | 0.20 |
ENSRNOT00000010555
|
C2cd4c
|
C2 calcium-dependent domain containing 4C |
| chr16_-_8564379 | 0.19 |
ENSRNOT00000060975
|
LOC680885
|
hypothetical protein LOC680885 |
| chr1_+_71416110 | 0.19 |
ENSRNOT00000084899
|
Zfp787
|
zinc finger protein 787 |
| chr12_+_18531990 | 0.19 |
ENSRNOT00000036606
|
Asmtl
|
acetylserotonin O-methyltransferase-like |
| chr12_-_13668515 | 0.19 |
ENSRNOT00000086847
|
Fscn1
|
fascin actin-bundling protein 1 |
| chr12_-_50383496 | 0.19 |
ENSRNOT00000000831
|
Tpst2
|
tyrosylprotein sulfotransferase 2 |
| chr14_-_1461543 | 0.19 |
ENSRNOT00000075656
|
Crlf2
|
cytokine receptor-like factor 2 |
| chr17_-_31411957 | 0.18 |
ENSRNOT00000060132
|
Psmg4
|
proteasome assembly chaperone 4 |
| chr12_-_18530724 | 0.18 |
ENSRNOT00000078999
|
Akap17a
|
A-kinase anchoring protein 17A |
| chr16_-_8685529 | 0.18 |
ENSRNOT00000092751
|
Slc18a3
|
solute carrier family 18 member A3 |
| chr1_-_83990588 | 0.18 |
ENSRNOT00000002052
|
Rab4b
|
RAB4B, member RAS oncogene family |
| chr17_+_9653561 | 0.18 |
ENSRNOT00000018899
|
Pdlim7
|
PDZ and LIM domain 7 |
| chr19_-_9931930 | 0.18 |
ENSRNOT00000085144
|
Ccdc113
|
coiled-coil domain containing 113 |
| chr1_-_216663720 | 0.18 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr1_+_266075782 | 0.18 |
ENSRNOT00000026471
|
Fbxl15
|
F-box and leucine-rich repeat protein 15 |
| chr1_-_101175570 | 0.18 |
ENSRNOT00000073918
|
Gfy
|
golgi-associated, olfactory signaling regulator |
| chr2_-_202816562 | 0.18 |
ENSRNOT00000020401
|
Fam46c
|
family with sequence similarity 46, member C |
| chr1_+_219345918 | 0.17 |
ENSRNOT00000025018
|
Cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr20_-_2701815 | 0.17 |
ENSRNOT00000061950
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr16_-_20890949 | 0.17 |
ENSRNOT00000081977
|
Homer3
|
homer scaffolding protein 3 |
| chr14_+_83752393 | 0.17 |
ENSRNOT00000081123
|
Selenom
|
selenoprotein M |
| chr10_-_91047177 | 0.17 |
ENSRNOT00000003986
|
C1ql1
|
complement C1q like 1 |
| chr3_+_13838304 | 0.17 |
ENSRNOT00000025067
|
Hspa5
|
heat shock protein family A member 5 |
| chr12_-_18540166 | 0.16 |
ENSRNOT00000001792
|
Il3ra
|
interleukin 3 receptor subunit alpha |
| chr3_+_171832500 | 0.16 |
ENSRNOT00000007554
|
Vapb
|
VAMP associated protein B and C |
| chr1_-_80544825 | 0.16 |
ENSRNOT00000057802
ENSRNOT00000040060 ENSRNOT00000067049 ENSRNOT00000052387 ENSRNOT00000073352 |
Relb
|
RELB proto-oncogene, NF-kB subunit |
| chr8_-_22874637 | 0.16 |
ENSRNOT00000064551
ENSRNOT00000090424 |
Dock6
|
dedicator of cytokinesis 6 |
| chr12_+_22229079 | 0.15 |
ENSRNOT00000001911
|
Gnb2
|
G protein subunit beta 2 |
| chr12_+_47218969 | 0.15 |
ENSRNOT00000081343
ENSRNOT00000038395 |
Unc119b
|
unc-119 lipid binding chaperone B |
| chr17_+_76002275 | 0.15 |
ENSRNOT00000092665
ENSRNOT00000086701 |
Echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr4_+_121612332 | 0.15 |
ENSRNOT00000077374
ENSRNOT00000084494 |
Txnrd3
|
thioredoxin reductase 3 |
| chr1_+_199655660 | 0.15 |
ENSRNOT00000027032
|
Armc5
|
armadillo repeat containing 5 |
| chr7_+_12619774 | 0.15 |
ENSRNOT00000015257
|
Med16
|
mediator complex subunit 16 |
| chr3_+_11921715 | 0.15 |
ENSRNOT00000021689
|
Fam129b
|
family with sequence similarity 129, member B |
| chr1_+_81474553 | 0.15 |
ENSRNOT00000083493
|
Phldb3
|
pleckstrin homology-like domain, family B, member 3 |
| chr12_-_4474729 | 0.15 |
ENSRNOT00000001418
|
Cers4
|
ceramide synthase 4 |
| chr17_+_31441630 | 0.15 |
ENSRNOT00000083705
ENSRNOT00000023582 |
Tubb2b
|
tubulin, beta 2B class IIb |
| chr1_-_88686615 | 0.14 |
ENSRNOT00000070921
|
Tbcb
|
tubulin folding cofactor B |
| chr10_+_106812739 | 0.14 |
ENSRNOT00000074225
|
Syngr2
|
synaptogyrin 2 |
| chr4_+_7662019 | 0.14 |
ENSRNOT00000014023
|
Fam126a
|
family with sequence similarity 126, member A |
| chr20_-_2701637 | 0.14 |
ENSRNOT00000049667
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr12_+_47551935 | 0.14 |
ENSRNOT00000056932
|
RGD1560398
|
RGD1560398 |
| chr5_-_173222440 | 0.14 |
ENSRNOT00000024897
|
Vwa1
|
von Willebrand factor A domain containing 1 |
| chr13_+_52588917 | 0.14 |
ENSRNOT00000011999
|
Phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr18_+_24570762 | 0.13 |
ENSRNOT00000021738
|
Polr2d
|
RNA polymerase II subunit D |
| chr19_+_10731855 | 0.13 |
ENSRNOT00000022277
|
Pllp
|
plasmolipin |
| chr1_-_101360971 | 0.13 |
ENSRNOT00000028164
|
Lin7b
|
lin-7 homolog B, crumbs cell polarity complex component |
| chr1_+_36320461 | 0.13 |
ENSRNOT00000023659
|
Srd5a1
|
steroid 5 alpha-reductase 1 |
| chr1_+_79831534 | 0.13 |
ENSRNOT00000057965
|
Nova2
|
NOVA alternative splicing regulator 2 |
| chr16_-_14348046 | 0.13 |
ENSRNOT00000018630
|
Cdhr1
|
cadherin-related family member 1 |
| chr20_-_4879779 | 0.13 |
ENSRNOT00000081924
|
Hspa1b
|
heat shock protein family A (Hsp70) member 1B |
| chr16_+_19051965 | 0.13 |
ENSRNOT00000016399
|
Slc35e1
|
solute carrier family 35, member E1 |
| chr1_+_101178104 | 0.13 |
ENSRNOT00000028072
|
Pth2
|
parathyroid hormone 2 |
| chr10_+_49299125 | 0.13 |
ENSRNOT00000074853
|
Tvp23b
|
trans-golgi network vesicle protein 23 homolog B |
| chrX_-_14890606 | 0.13 |
ENSRNOT00000049864
|
RGD1560784
|
similar to RIKEN cDNA B630019K06 |
| chr16_-_19894591 | 0.13 |
ENSRNOT00000085940
|
Ano8
|
anoctamin 8 |
| chr19_+_57047830 | 0.13 |
ENSRNOT00000080860
|
Galnt2
|
polypeptide N-acetylgalactosaminyltransferase 2 |
| chr12_+_48481750 | 0.13 |
ENSRNOT00000000886
|
Coro1c
|
coronin 1C |
| chr1_+_261229347 | 0.13 |
ENSRNOT00000018485
|
Ubtd1
|
ubiquitin domain containing 1 |
| chr1_-_80580662 | 0.13 |
ENSRNOT00000024779
|
Clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr3_-_82368357 | 0.13 |
ENSRNOT00000000052
|
Cd82
|
Cd82 molecule |
| chr12_-_38995570 | 0.13 |
ENSRNOT00000001806
|
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chr8_+_22050222 | 0.12 |
ENSRNOT00000028096
|
Icam5
|
intercellular adhesion molecule 5 |
| chr10_+_55940533 | 0.12 |
ENSRNOT00000012061
|
RGD1563441
|
similar to RIKEN cDNA A030009H04 |
| chr3_+_171037957 | 0.12 |
ENSRNOT00000008764
|
Rbm38
|
RNA binding motif protein 38 |
| chr1_+_101412736 | 0.12 |
ENSRNOT00000067468
|
Lhb
|
luteinizing hormone beta polypeptide |
| chr2_-_195907450 | 0.12 |
ENSRNOT00000089402
|
Tuft1
|
tuftelin 1 |
| chr4_+_171748273 | 0.12 |
ENSRNOT00000009998
|
Dera
|
deoxyribose-phosphate aldolase |
| chr2_-_195908037 | 0.12 |
ENSRNOT00000079776
|
Tuft1
|
tuftelin 1 |
| chr20_+_5050327 | 0.12 |
ENSRNOT00000083353
|
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr16_-_7758189 | 0.12 |
ENSRNOT00000026588
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr7_+_11992543 | 0.12 |
ENSRNOT00000024576
|
Abhd17a
|
abhydrolase domain containing 17A |
| chr5_-_136706936 | 0.12 |
ENSRNOT00000081925
ENSRNOT00000068130 |
Ccdc24
|
coiled-coil domain containing 24 |
| chr5_-_136002900 | 0.12 |
ENSRNOT00000025197
|
Plk3
|
polo-like kinase 3 |
| chr4_+_180961142 | 0.12 |
ENSRNOT00000002500
|
Med21
|
mediator complex subunit 21 |
| chr5_+_157642751 | 0.12 |
ENSRNOT00000083577
ENSRNOT00000010308 |
Capzb
|
capping actin protein of muscle Z-line beta subunit |
| chr19_-_25961666 | 0.12 |
ENSRNOT00000004091
|
Calr
|
calreticulin |
| chr16_+_83358116 | 0.12 |
ENSRNOT00000031109
|
Rab20
|
RAB20, member RAS oncogene family |
| chr19_+_20607507 | 0.12 |
ENSRNOT00000000011
|
Cbln1
|
cerebellin 1 precursor |
| chr6_-_109004598 | 0.12 |
ENSRNOT00000007790
|
Pgf
|
placental growth factor |
| chr9_-_10047507 | 0.12 |
ENSRNOT00000072118
|
Alkbh7
|
alkB homolog 7 |
| chr4_-_179477070 | 0.12 |
ENSRNOT00000030680
|
Casc1
|
cancer susceptibility candidate 1 |
| chr5_-_153790157 | 0.12 |
ENSRNOT00000025051
|
Rcan3
|
RCAN family member 3 |
| chr17_+_78817529 | 0.12 |
ENSRNOT00000021918
|
Meig1
|
meiosis/spermiogenesis associated 1 |
| chr10_-_91117889 | 0.12 |
ENSRNOT00000031498
|
Dcakd
|
dephospho-CoA kinase domain containing |
| chr3_-_111080705 | 0.12 |
ENSRNOT00000079339
|
Rhov
|
ras homolog family member V |
| chr10_-_56403188 | 0.12 |
ENSRNOT00000019947
|
Chrnb1
|
cholinergic receptor nicotinic beta 1 subunit |
| chr1_-_91074294 | 0.12 |
ENSRNOT00000075236
|
LOC687508
|
similar to Cytochrome c oxidase polypeptide VIIa-heart, mitochondrial precursor (Cytochrome c oxidase subunit VIIa-H) (COX VIIa-M) |
| chr1_-_64099277 | 0.11 |
ENSRNOT00000084846
|
Tsen34
|
tRNA splicing endonuclease subunit 34 |
| chr7_+_143629455 | 0.11 |
ENSRNOT00000073951
|
Krt18
|
keratin 18 |
| chr3_+_164424515 | 0.11 |
ENSRNOT00000083876
|
Cebpb
|
CCAAT/enhancer binding protein beta |
| chr10_+_7041510 | 0.11 |
ENSRNOT00000003514
|
Carhsp1
|
calcium regulated heat stable protein 1 |
| chr16_+_19767264 | 0.11 |
ENSRNOT00000051802
ENSRNOT00000092073 |
Ocel1
|
occludin/ELL domain containing 1 |
| chr3_+_151553578 | 0.11 |
ENSRNOT00000045484
|
Ergic3
|
ERGIC and golgi 3 |
| chr12_+_40619377 | 0.11 |
ENSRNOT00000001822
ENSRNOT00000081016 |
Erp29
|
endoplasmic reticulum protein 29 |
| chr8_+_96564877 | 0.11 |
ENSRNOT00000017879
|
Mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr1_-_190965115 | 0.11 |
ENSRNOT00000023483
|
AC103221.1
|
|
| chr18_+_79406381 | 0.11 |
ENSRNOT00000022303
ENSRNOT00000058295 ENSRNOT00000058296 ENSRNOT00000022280 |
Mbp
|
myelin basic protein |
| chr8_+_119382011 | 0.11 |
ENSRNOT00000056060
|
Lrrfip2
|
LRR binding FLII interacting protein 2 |
| chr3_+_15379109 | 0.11 |
ENSRNOT00000036495
|
Morn5
|
MORN repeat containing 5 |
| chr20_+_13156241 | 0.11 |
ENSRNOT00000050531
|
Prmt2
|
protein arginine methyltransferase 2 |
| chr10_-_46593009 | 0.11 |
ENSRNOT00000047053
|
Srebf1
|
sterol regulatory element binding transcription factor 1 |
| chr7_+_11215328 | 0.11 |
ENSRNOT00000061194
|
LOC690617
|
hypothetical protein LOC690617 |
| chr8_+_53678777 | 0.11 |
ENSRNOT00000045944
|
Drd2
|
dopamine receptor D2 |
| chr1_+_274245184 | 0.10 |
ENSRNOT00000018889
|
Dusp5
|
dual specificity phosphatase 5 |
| chr5_+_157282669 | 0.10 |
ENSRNOT00000022827
|
Pla2g2a
|
phospholipase A2 group IIA |
| chr19_+_26106838 | 0.10 |
ENSRNOT00000035987
|
Hook2
|
hook microtubule-tethering protein 2 |
| chr1_+_88182585 | 0.10 |
ENSRNOT00000044145
|
Yif1b
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chr3_+_150323116 | 0.10 |
ENSRNOT00000023429
ENSRNOT00000080485 |
Raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr19_-_19509087 | 0.10 |
ENSRNOT00000019301
|
Nkd1
|
naked cuticle homolog 1 |
| chr7_+_12314848 | 0.10 |
ENSRNOT00000028969
|
Gamt
|
guanidinoacetate N-methyltransferase |
| chr10_-_102200596 | 0.10 |
ENSRNOT00000081519
|
Fam104a
|
family with sequence similarity 104, member A |
| chr5_+_159845774 | 0.10 |
ENSRNOT00000012328
|
Epha2
|
Eph receptor A2 |
| chr12_-_11265865 | 0.10 |
ENSRNOT00000001315
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr2_+_187737770 | 0.10 |
ENSRNOT00000036143
ENSRNOT00000092611 ENSRNOT00000092620 |
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr1_-_221281180 | 0.10 |
ENSRNOT00000028379
|
Cdc42ep2
|
CDC42 effector protein 2 |
| chr1_+_91042635 | 0.10 |
ENSRNOT00000028211
|
LOC103690005
|
tubulin-folding cofactor B |
| chr17_+_54280851 | 0.10 |
ENSRNOT00000024022
|
Arhgap12
|
Rho GTPase activating protein 12 |
| chr3_+_151032952 | 0.10 |
ENSRNOT00000064013
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr10_-_55642681 | 0.10 |
ENSRNOT00000057157
|
AC129753.1
|
|
| chr14_-_2297155 | 0.10 |
ENSRNOT00000000071
|
Pcgf3
|
polycomb group ring finger 3 |
| chr1_+_221448661 | 0.10 |
ENSRNOT00000072493
|
LOC100910252
|
sororin-like |
| chr1_+_198690794 | 0.10 |
ENSRNOT00000023999
|
Zfp771
|
zinc finger protein 771 |
| chr1_-_89329185 | 0.10 |
ENSRNOT00000078284
|
Cd22
|
CD22 molecule |
| chr19_-_52499433 | 0.10 |
ENSRNOT00000021954
|
Cotl1
|
coactosin-like F-actin binding protein 1 |
| chr16_-_75648538 | 0.09 |
ENSRNOT00000048414
|
Defb14
|
defensin beta 14 |
| chr4_-_113610243 | 0.09 |
ENSRNOT00000008813
|
Hk2
|
hexokinase 2 |
| chr2_+_251453644 | 0.09 |
ENSRNOT00000041897
|
Znhit6
|
zinc finger, HIT-type containing 6 |
| chr16_-_19942343 | 0.09 |
ENSRNOT00000087162
ENSRNOT00000091906 |
Bst2
|
bone marrow stromal cell antigen 2 |
| chr10_-_45543897 | 0.09 |
ENSRNOT00000003926
|
Guk1
|
guanylate kinase 1 |
| chr12_+_22165486 | 0.09 |
ENSRNOT00000001890
|
Mospd3
|
motile sperm domain containing 3 |
| chr2_+_190073815 | 0.09 |
ENSRNOT00000015473
|
S100a8
|
S100 calcium binding protein A8 |
| chrX_-_113584459 | 0.09 |
ENSRNOT00000025923
|
Kcne5
|
potassium voltage-gated channel subfamily E regulatory subunit 5 |
| chr1_+_218632764 | 0.09 |
ENSRNOT00000019861
|
Tesmin
|
testis expressed metallothionein like protein |
| chr3_-_11410732 | 0.09 |
ENSRNOT00000034930
|
RGD1561113
|
similar to Hypothetical UPF0184 protein C9orf16 homolog |
| chr7_+_11545024 | 0.09 |
ENSRNOT00000073221
|
Slc39a3
|
solute carrier family 39 member 3 |
| chr19_+_55094585 | 0.09 |
ENSRNOT00000068452
|
Zfpm1
|
zinc finger protein, multitype 1 |
| chr16_+_21288876 | 0.09 |
ENSRNOT00000027990
|
Cilp2
|
cartilage intermediate layer protein 2 |
| chr6_-_6842758 | 0.09 |
ENSRNOT00000006094
|
Kcng3
|
potassium voltage-gated channel modifier subfamily G member 3 |
| chr4_+_145580799 | 0.09 |
ENSRNOT00000013727
|
Vhl
|
von Hippel-Lindau tumor suppressor |
| chr7_+_12471824 | 0.09 |
ENSRNOT00000068197
|
Sbno2
|
strawberry notch homolog 2 |
| chr9_-_3965939 | 0.09 |
ENSRNOT00000088289
|
AABR07066144.1
|
|
| chr1_+_72732668 | 0.09 |
ENSRNOT00000024251
|
Hspbp1
|
HSPA (heat shock 70) binding protein, cytoplasmic cochaperone 1 |
| chr1_+_282134981 | 0.09 |
ENSRNOT00000036203
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr4_-_100883038 | 0.09 |
ENSRNOT00000041880
|
LOC100364435
|
thymosin, beta 10-like |
| chr9_+_113699170 | 0.09 |
ENSRNOT00000017915
|
Twsg1
|
twisted gastrulation BMP signaling modulator 1 |
| chr12_+_47074200 | 0.09 |
ENSRNOT00000014910
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr19_+_24747178 | 0.09 |
ENSRNOT00000038879
ENSRNOT00000079106 |
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr12_+_47698947 | 0.09 |
ENSRNOT00000001586
|
Trpv4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chr7_-_70842405 | 0.08 |
ENSRNOT00000047449
|
Nxph4
|
neurexophilin 4 |
| chr4_-_158010166 | 0.08 |
ENSRNOT00000026633
|
Cd9
|
CD9 molecule |
| chr17_-_66397653 | 0.08 |
ENSRNOT00000024098
|
Actn2
|
actinin alpha 2 |
| chrX_+_29525256 | 0.08 |
ENSRNOT00000050018
|
Rab9a
|
RAB9A, member RAS oncogene family |
| chr12_-_13210758 | 0.08 |
ENSRNOT00000001438
|
Kdelr2
|
KDEL endoplasmic reticulum protein retention receptor 2 |
| chr1_-_88112683 | 0.08 |
ENSRNOT00000090615
|
Spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr3_-_147809919 | 0.08 |
ENSRNOT00000085849
|
Rbck1
|
RANBP2-type and C3HC4-type zinc finger containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf12_Klf14_Sp4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.4 | GO:0031439 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
| 0.1 | 0.4 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.4 | GO:1902568 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.3 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 0.3 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.2 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.2 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.2 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.1 | 0.2 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.6 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 0.2 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.2 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 0.1 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.1 | 0.2 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 0.2 | GO:0015746 | citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.6 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 0.0 | 0.1 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 0.0 | 0.0 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.0 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.4 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0009397 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.3 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 0.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0019541 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.2 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.2 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0072566 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.1 | GO:1903002 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.1 | GO:1901491 | notochord formation(GO:0014028) negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.1 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0052428 | negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.2 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:1901423 | response to benzene(GO:1901423) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.1 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.6 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.2 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0071865 | mammary gland bud morphogenesis(GO:0060648) regulation of apoptotic process in bone marrow(GO:0071865) negative regulation of apoptotic process in bone marrow(GO:0071866) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.3 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.0 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.0 | 0.1 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.0 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.0 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.0 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.2 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.0 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.0 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.0 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) histidine catabolic process(GO:0006548) |
| 0.0 | 0.0 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) |
| 0.0 | 0.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.0 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.0 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.1 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.0 | GO:1901093 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.0 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.0 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.0 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.0 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.0 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.1 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.0 | 0.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.0 | GO:1904528 | positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.2 | GO:0034030 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:1904779 | regulation of protein localization to centrosome(GO:1904779) |
| 0.0 | 0.1 | GO:0006702 | androgen biosynthetic process(GO:0006702) circadian sleep/wake cycle, REM sleep(GO:0042747) |
| 0.0 | 0.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.0 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.3 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.6 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.2 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.0 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.0 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.0 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.0 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.0 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.4 | GO:0031249 | denatured protein binding(GO:0031249) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.5 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.4 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.0 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.0 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 0.0 | GO:0030283 | testosterone dehydrogenase [NAD(P)] activity(GO:0030283) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 0.0 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.0 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0046979 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.0 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |