Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
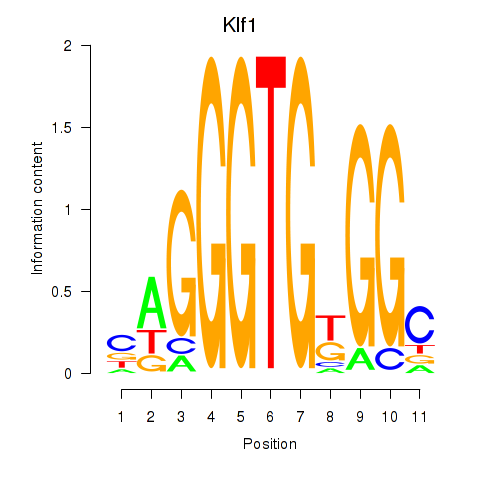
Results for Klf1
Z-value: 0.57
Transcription factors associated with Klf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf1
|
ENSRNOG00000003443 | Kruppel like factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf1 | rn6_v1_chr19_+_26016382_26016382 | 0.62 | 2.7e-01 | Click! |
Activity profile of Klf1 motif
Sorted Z-values of Klf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_77632628 | 0.37 |
ENSRNOT00000073915
|
Basp1
|
brain abundant, membrane attached signal protein 1 |
| chr1_+_168964202 | 0.36 |
ENSRNOT00000089102
|
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr10_-_15590220 | 0.36 |
ENSRNOT00000048977
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr10_-_56962161 | 0.32 |
ENSRNOT00000026038
|
Alox15
|
arachidonate 15-lipoxygenase |
| chr9_+_82647071 | 0.31 |
ENSRNOT00000027135
|
Asic4
|
acid sensing ion channel subunit family member 4 |
| chr10_-_15577977 | 0.29 |
ENSRNOT00000052292
|
Hba-a3
|
hemoglobin alpha, adult chain 3 |
| chr1_+_168945449 | 0.29 |
ENSRNOT00000087661
ENSRNOT00000019913 |
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr4_-_100883038 | 0.28 |
ENSRNOT00000041880
|
LOC100364435
|
thymosin, beta 10-like |
| chr4_-_100883275 | 0.25 |
ENSRNOT00000022846
|
LOC100364435
|
thymosin, beta 10-like |
| chr1_+_198690794 | 0.25 |
ENSRNOT00000023999
|
Zfp771
|
zinc finger protein 771 |
| chr1_+_168957460 | 0.25 |
ENSRNOT00000090745
|
LOC103694857
|
hemoglobin subunit beta-2 |
| chr1_-_78212350 | 0.24 |
ENSRNOT00000071098
|
Inafm1
|
InaF-motif containing 1 |
| chr1_-_168972725 | 0.23 |
ENSRNOT00000090422
|
Hbb
|
hemoglobin subunit beta |
| chr10_-_40375605 | 0.22 |
ENSRNOT00000014464
|
Anxa6
|
annexin A6 |
| chr17_-_9695292 | 0.21 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr11_+_30904733 | 0.21 |
ENSRNOT00000036027
|
Mrap
|
melanocortin 2 receptor accessory protein |
| chr6_+_135856218 | 0.19 |
ENSRNOT00000066715
|
RGD1560608
|
similar to novel protein |
| chr3_+_154395187 | 0.18 |
ENSRNOT00000050810
|
Vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr2_-_30127269 | 0.17 |
ENSRNOT00000023869
|
Cartpt
|
CART prepropeptide |
| chr1_-_47331412 | 0.17 |
ENSRNOT00000046746
|
Ezr
|
ezrin |
| chr6_-_1942972 | 0.16 |
ENSRNOT00000048711
|
Cdc42ep3
|
CDC42 effector protein 3 |
| chr5_-_136762986 | 0.15 |
ENSRNOT00000026852
|
Artn
|
artemin |
| chr12_-_30304036 | 0.15 |
ENSRNOT00000001218
|
Nupr2
|
nuclear protein 2, transcriptional regulator |
| chr1_+_80321585 | 0.15 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr20_-_5618254 | 0.15 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr3_+_11679530 | 0.15 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr10_+_86950557 | 0.15 |
ENSRNOT00000014153
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr20_+_5050327 | 0.15 |
ENSRNOT00000083353
|
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr5_+_157282669 | 0.14 |
ENSRNOT00000022827
|
Pla2g2a
|
phospholipase A2 group IIA |
| chr12_+_22641104 | 0.14 |
ENSRNOT00000001916
|
Serpine1
|
serpin family E member 1 |
| chr1_-_82610350 | 0.14 |
ENSRNOT00000028177
|
Cyp2s1
|
cytochrome P450, family 2, subfamily s, polypeptide 1 |
| chr5_-_72287669 | 0.14 |
ENSRNOT00000022255
|
Klf4
|
Kruppel like factor 4 |
| chr5_+_151413382 | 0.14 |
ENSRNOT00000012626
|
Cd164l2
|
CD164 molecule like 2 |
| chr8_+_44847157 | 0.13 |
ENSRNOT00000080288
|
Clmp
|
CXADR-like membrane protein |
| chr18_+_32594958 | 0.13 |
ENSRNOT00000018511
|
Spry4
|
sprouty RTK signaling antagonist 4 |
| chr5_-_173222440 | 0.13 |
ENSRNOT00000024897
|
Vwa1
|
von Willebrand factor A domain containing 1 |
| chr1_+_81230612 | 0.12 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr19_+_56220755 | 0.12 |
ENSRNOT00000023452
|
Tubb3
|
tubulin, beta 3 class III |
| chr5_-_152324469 | 0.12 |
ENSRNOT00000020688
|
Cd52
|
CD52 molecule |
| chr6_+_8886591 | 0.12 |
ENSRNOT00000091510
ENSRNOT00000089174 |
Six3
|
SIX homeobox 3 |
| chr3_+_66673246 | 0.12 |
ENSRNOT00000081338
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr1_-_198559568 | 0.12 |
ENSRNOT00000023080
|
Qprt
|
quinolinate phosphoribosyltransferase |
| chr8_+_127144903 | 0.12 |
ENSRNOT00000013530
|
Eomes
|
eomesodermin |
| chr3_-_72289310 | 0.12 |
ENSRNOT00000038250
|
Rtn4rl2
|
reticulon 4 receptor-like 2 |
| chr2_+_206064394 | 0.12 |
ENSRNOT00000077739
|
Syt6
|
synaptotagmin 6 |
| chr3_-_177201525 | 0.12 |
ENSRNOT00000022451
|
Rgs19
|
regulator of G-protein signaling 19 |
| chr16_-_14300951 | 0.12 |
ENSRNOT00000017038
|
Rgr
|
retinal G protein coupled receptor |
| chr4_-_157439507 | 0.12 |
ENSRNOT00000081802
|
Ptms
|
parathymosin |
| chr10_-_91047177 | 0.12 |
ENSRNOT00000003986
|
C1ql1
|
complement C1q like 1 |
| chr12_+_24651314 | 0.11 |
ENSRNOT00000077016
ENSRNOT00000071569 |
Vps37d
|
VPS37D, ESCRT-I subunit |
| chr2_-_202816562 | 0.11 |
ENSRNOT00000020401
|
Fam46c
|
family with sequence similarity 46, member C |
| chr1_+_242572533 | 0.11 |
ENSRNOT00000035123
|
Tmem252
|
transmembrane protein 252 |
| chr1_-_164307084 | 0.11 |
ENSRNOT00000086091
|
Serpinh1
|
serpin family H member 1 |
| chr4_-_28702559 | 0.11 |
ENSRNOT00000013910
ENSRNOT00000013937 |
Calcr
|
calcitonin receptor |
| chr20_+_5049496 | 0.11 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr17_+_9653561 | 0.11 |
ENSRNOT00000018899
|
Pdlim7
|
PDZ and LIM domain 7 |
| chr14_-_37770059 | 0.11 |
ENSRNOT00000029721
|
Slc10a4
|
solute carrier family 10, member 4 |
| chr3_-_67668772 | 0.11 |
ENSRNOT00000010247
|
Frzb
|
frizzled-related protein |
| chr13_-_80738634 | 0.11 |
ENSRNOT00000081551
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr4_+_171250818 | 0.11 |
ENSRNOT00000040576
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr20_+_5040337 | 0.10 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr7_+_12974169 | 0.10 |
ENSRNOT00000010555
|
C2cd4c
|
C2 calcium-dependent domain containing 4C |
| chr1_+_83003841 | 0.10 |
ENSRNOT00000057384
|
Ceacam4
|
carcinoembryonic antigen-related cell adhesion molecule 4 |
| chr11_+_88122271 | 0.10 |
ENSRNOT00000002540
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr19_+_23389375 | 0.10 |
ENSRNOT00000018629
|
Sall1
|
spalt-like transcription factor 1 |
| chr5_+_147714163 | 0.10 |
ENSRNOT00000012663
|
Marcksl1
|
MARCKS-like 1 |
| chr3_-_172537877 | 0.10 |
ENSRNOT00000072069
|
Ctsz
|
cathepsin Z |
| chr6_+_29977797 | 0.10 |
ENSRNOT00000071784
|
Fkbp1b
|
FK506 binding protein 1B |
| chr1_+_89215266 | 0.10 |
ENSRNOT00000093612
ENSRNOT00000084799 |
Dmkn
|
dermokine |
| chr1_-_101360971 | 0.10 |
ENSRNOT00000028164
|
Lin7b
|
lin-7 homolog B, crumbs cell polarity complex component |
| chr1_-_84812486 | 0.10 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chr13_+_110743098 | 0.10 |
ENSRNOT00000075086
|
Rd3
|
retinal degeneration 3 |
| chr14_-_82055290 | 0.10 |
ENSRNOT00000058062
|
RGD1560394
|
RGD1560394 |
| chr4_-_14490446 | 0.10 |
ENSRNOT00000009132
|
Sema3c
|
semaphorin 3C |
| chr13_-_32427177 | 0.10 |
ENSRNOT00000044628
|
Cdh19
|
cadherin 19 |
| chr11_+_87549059 | 0.10 |
ENSRNOT00000002567
|
Ccdc74a
|
coiled-coil domain containing 74A |
| chr6_-_46631983 | 0.10 |
ENSRNOT00000045963
|
Sox11
|
SRY box 11 |
| chr1_-_89369960 | 0.09 |
ENSRNOT00000028545
|
Hamp
|
hepcidin antimicrobial peptide |
| chr17_+_31441630 | 0.09 |
ENSRNOT00000083705
ENSRNOT00000023582 |
Tubb2b
|
tubulin, beta 2B class IIb |
| chr9_+_81518584 | 0.09 |
ENSRNOT00000084309
|
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr1_+_267607416 | 0.09 |
ENSRNOT00000087894
ENSRNOT00000076367 ENSRNOT00000016851 |
Gsto2
Gsto1
|
glutathione S-transferase omega 2 glutathione S-transferase omega 1 |
| chr19_+_37235001 | 0.09 |
ENSRNOT00000020908
|
Nol3
|
nucleolar protein 3 |
| chr3_+_81294275 | 0.09 |
ENSRNOT00000009068
|
RGD1563263
|
similar to RIKEN cDNA 1700029I15 |
| chrX_-_106558366 | 0.09 |
ENSRNOT00000042126
|
Bex2
|
brain expressed X-linked 2 |
| chr7_+_142912316 | 0.09 |
ENSRNOT00000010171
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr15_+_37790141 | 0.09 |
ENSRNOT00000076392
ENSRNOT00000091953 |
Il17d
|
interleukin 17D |
| chr10_-_14022452 | 0.09 |
ENSRNOT00000016554
|
Npw
|
neuropeptide W |
| chr20_+_8165307 | 0.09 |
ENSRNOT00000000637
|
Pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr2_-_195908037 | 0.09 |
ENSRNOT00000079776
|
Tuft1
|
tuftelin 1 |
| chr1_-_265506046 | 0.09 |
ENSRNOT00000023963
|
Npm3
|
nucleophosmin/nucleoplasmin, 3 |
| chr15_-_33193537 | 0.09 |
ENSRNOT00000016401
|
Haus4
|
HAUS augmin-like complex, subunit 4 |
| chr1_-_101883744 | 0.09 |
ENSRNOT00000028635
|
Syngr4
|
synaptogyrin 4 |
| chr7_+_139762614 | 0.09 |
ENSRNOT00000031157
|
Ccdc184
|
coiled-coil domain containing 184 |
| chr7_-_121232741 | 0.09 |
ENSRNOT00000023196
|
Pdgfb
|
platelet derived growth factor subunit B |
| chr20_+_3178250 | 0.09 |
ENSRNOT00000082116
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr4_-_114853868 | 0.09 |
ENSRNOT00000090828
|
Wdr54
|
WD repeat domain 54 |
| chr3_+_147226004 | 0.09 |
ENSRNOT00000012765
|
Tmem74b
|
transmembrane protein 74B |
| chr1_+_88182585 | 0.08 |
ENSRNOT00000044145
|
Yif1b
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chr1_-_101175570 | 0.08 |
ENSRNOT00000073918
|
Gfy
|
golgi-associated, olfactory signaling regulator |
| chr11_+_33845463 | 0.08 |
ENSRNOT00000041838
|
Cbr1
|
carbonyl reductase 1 |
| chr3_-_123702732 | 0.08 |
ENSRNOT00000028859
|
RGD1311739
|
similar to RIKEN cDNA 1700037H04 |
| chr1_-_261446570 | 0.08 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr12_-_6703979 | 0.08 |
ENSRNOT00000061246
|
Tex26
|
testis expressed 26 |
| chr20_-_14620019 | 0.08 |
ENSRNOT00000001779
|
Gnaz
|
G protein subunit alpha z |
| chr10_+_48903540 | 0.08 |
ENSRNOT00000004248
|
Trpv2
|
transient receptor potential cation channel, subfamily V, member 2 |
| chr1_-_216663720 | 0.08 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr1_+_140602542 | 0.08 |
ENSRNOT00000085570
|
Isg20
|
interferon stimulated exonuclease gene 20 |
| chr8_-_13606040 | 0.08 |
ENSRNOT00000013577
ENSRNOT00000083395 |
Panx1
|
Pannexin 1 |
| chr4_+_168832910 | 0.08 |
ENSRNOT00000011134
|
Gprc5a
|
G protein-coupled receptor, class C, group 5, member A |
| chr11_+_33812989 | 0.08 |
ENSRNOT00000042283
ENSRNOT00000075985 |
Cbr1
|
carbonyl reductase 1 |
| chr1_+_282134981 | 0.08 |
ENSRNOT00000036203
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr10_-_105668593 | 0.08 |
ENSRNOT00000016622
|
St6galnac2
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
| chr4_+_114854458 | 0.08 |
ENSRNOT00000013312
|
LOC500227
|
hypothetical gene supported by BC079424 |
| chr1_+_276659542 | 0.08 |
ENSRNOT00000019681
|
Tcf7l2
|
transcription factor 7 like 2 |
| chr2_+_22909569 | 0.08 |
ENSRNOT00000073871
|
Homer1
|
homer scaffolding protein 1 |
| chr1_-_89473904 | 0.08 |
ENSRNOT00000089474
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr14_+_10692764 | 0.08 |
ENSRNOT00000003012
|
LOC100910270
|
uncharacterized LOC100910270 |
| chr13_-_112005052 | 0.08 |
ENSRNOT00000007879
|
G0s2
|
G0/G1switch 2 |
| chr12_-_19599374 | 0.08 |
ENSRNOT00000001849
|
Gpc2
|
glypican 2 |
| chr13_-_52197205 | 0.08 |
ENSRNOT00000009712
|
Shisa4
|
shisa family member 4 |
| chr6_+_132246602 | 0.08 |
ENSRNOT00000009896
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr6_-_135939534 | 0.08 |
ENSRNOT00000052237
|
Diras3
|
DIRAS family GTPase 3 |
| chr2_+_211176556 | 0.08 |
ENSRNOT00000055880
|
Psrc1
|
proline and serine rich coiled-coil 1 |
| chr10_-_108473377 | 0.08 |
ENSRNOT00000070934
|
Sgsh
|
N-sulfoglucosamine sulfohydrolase |
| chr1_-_78333321 | 0.08 |
ENSRNOT00000020402
|
Sae1
|
SUMO1 activating enzyme subunit 1 |
| chr17_+_15814132 | 0.08 |
ENSRNOT00000032997
|
Susd3
|
sushi domain containing 3 |
| chr5_+_147476221 | 0.08 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr10_+_7041510 | 0.08 |
ENSRNOT00000003514
|
Carhsp1
|
calcium regulated heat stable protein 1 |
| chr9_-_94250809 | 0.08 |
ENSRNOT00000026387
|
Ecel1
|
endothelin converting enzyme-like 1 |
| chr14_-_107617161 | 0.08 |
ENSRNOT00000012286
|
B3gnt2
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
| chr2_+_189922996 | 0.08 |
ENSRNOT00000016499
|
S100a16
|
S100 calcium binding protein A16 |
| chr12_+_47074200 | 0.08 |
ENSRNOT00000014910
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr1_+_192613372 | 0.08 |
ENSRNOT00000016632
|
Cacng3
|
calcium voltage-gated channel auxiliary subunit gamma 3 |
| chr20_-_29511382 | 0.07 |
ENSRNOT00000085026
|
Ddit4
|
DNA-damage-inducible transcript 4 |
| chrX_+_68782872 | 0.07 |
ENSRNOT00000075995
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr20_-_14282873 | 0.07 |
ENSRNOT00000001759
|
Adora2a
|
adenosine A2a receptor |
| chr8_+_75687100 | 0.07 |
ENSRNOT00000038677
|
Anxa2
|
annexin A2 |
| chr8_+_99977334 | 0.07 |
ENSRNOT00000085808
ENSRNOT00000056704 ENSRNOT00000041859 |
Plod2
|
procollagen lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr5_+_140923914 | 0.07 |
ENSRNOT00000020929
|
Heyl
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr6_+_137164535 | 0.07 |
ENSRNOT00000018225
|
Inf2
|
inverted formin, FH2 and WH2 domain containing |
| chr1_-_88111293 | 0.07 |
ENSRNOT00000077195
|
Spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr5_-_152358643 | 0.07 |
ENSRNOT00000021734
|
Sh3bgrl3
|
SH3 domain binding glutamate-rich protein like 3 |
| chr13_-_91228901 | 0.07 |
ENSRNOT00000071728
ENSRNOT00000073643 ENSRNOT00000071897 |
LOC100911825
|
low affinity immunoglobulin gamma Fc region receptor III-like |
| chr1_-_226501920 | 0.07 |
ENSRNOT00000050716
|
Lrrc10b
|
leucine rich repeat containing 10B |
| chr12_+_2534212 | 0.07 |
ENSRNOT00000001399
|
Ctxn1
|
cortexin 1 |
| chr7_-_145154131 | 0.07 |
ENSRNOT00000055271
|
Ppp1r1a
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr7_-_124367630 | 0.07 |
ENSRNOT00000055978
|
Ttll1
|
tubulin tyrosine ligase like 1 |
| chr10_-_89374516 | 0.07 |
ENSRNOT00000028084
|
Vat1
|
vesicle amine transport 1 |
| chr10_-_50402616 | 0.07 |
ENSRNOT00000004546
|
Hs3st3b1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
| chr4_-_170763916 | 0.07 |
ENSRNOT00000071512
|
Hist1h4m
|
histone cluster 1, H4m |
| chr8_-_70436028 | 0.07 |
ENSRNOT00000077152
|
Slc24a1
|
solute carrier family 24 member 1 |
| chr3_-_10226286 | 0.07 |
ENSRNOT00000093627
|
Fubp3
|
far upstream element binding protein 3 |
| chr1_-_214252456 | 0.07 |
ENSRNOT00000023504
|
Irf7
|
interferon regulatory factor 7 |
| chr16_-_71809925 | 0.07 |
ENSRNOT00000022425
|
Tm2d2
|
TM2 domain containing 2 |
| chr9_-_43127887 | 0.07 |
ENSRNOT00000021685
|
Ankrd39
|
ankyrin repeat domain 39 |
| chr1_+_85213652 | 0.07 |
ENSRNOT00000092044
|
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr19_-_6293 | 0.07 |
ENSRNOT00000074240
|
LOC679149
|
similar to carboxylesterase 2 (intestine, liver) |
| chr11_+_31428358 | 0.07 |
ENSRNOT00000002827
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr1_+_81208212 | 0.07 |
ENSRNOT00000026288
|
Lypd5
|
Ly6/Plaur domain containing 5 |
| chr1_-_88686615 | 0.07 |
ENSRNOT00000070921
|
Tbcb
|
tubulin folding cofactor B |
| chr3_+_66673071 | 0.07 |
ENSRNOT00000034769
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr3_+_110466790 | 0.07 |
ENSRNOT00000089434
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr7_-_70348780 | 0.07 |
ENSRNOT00000072468
|
March9
|
membrane associated ring-CH-type finger 9 |
| chr5_+_152681101 | 0.07 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chrX_+_15273933 | 0.07 |
ENSRNOT00000075082
|
LOC108348091
|
erythroid transcription factor |
| chr4_+_55715744 | 0.07 |
ENSRNOT00000010429
|
Arf5
|
ADP-ribosylation factor 5 |
| chr17_+_76002275 | 0.07 |
ENSRNOT00000092665
ENSRNOT00000086701 |
Echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr3_-_170955399 | 0.07 |
ENSRNOT00000084990
|
Bmp7
|
bone morphogenetic protein 7 |
| chr20_-_4921348 | 0.07 |
ENSRNOT00000082497
ENSRNOT00000041151 |
RT1-CE4
|
RT1 class I, locus CE4 |
| chrX_+_65226748 | 0.07 |
ENSRNOT00000076181
|
Msn
|
moesin |
| chr7_-_18683440 | 0.07 |
ENSRNOT00000068323
|
Rps28
|
ribosomal protein S28 |
| chr20_+_5351605 | 0.07 |
ENSRNOT00000089306
ENSRNOT00000041590 ENSRNOT00000081240 ENSRNOT00000082538 |
RT1-A1
|
RT1 class Ia, locus A1 |
| chr10_-_56531483 | 0.07 |
ENSRNOT00000022343
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr3_-_176144531 | 0.07 |
ENSRNOT00000082266
|
Tcfl5
|
transcription factor like 5 |
| chr10_-_40381886 | 0.07 |
ENSRNOT00000050213
|
LOC100912427
|
nuclease-sensitive element-binding protein 1-like |
| chr9_-_88357182 | 0.06 |
ENSRNOT00000041176
|
Col4a4
|
collagen type IV alpha 4 chain |
| chr10_-_56429748 | 0.06 |
ENSRNOT00000020675
ENSRNOT00000092704 |
Spem1
|
spermatid maturation 1 |
| chr7_-_114380613 | 0.06 |
ENSRNOT00000011898
|
Ago2
|
argonaute 2, RISC catalytic component |
| chr6_-_43862131 | 0.06 |
ENSRNOT00000089859
|
Cys1
|
cystin 1 |
| chr12_+_17736287 | 0.06 |
ENSRNOT00000091476
|
Pdgfa
|
platelet derived growth factor subunit A |
| chr10_+_12994683 | 0.06 |
ENSRNOT00000004702
|
Hcfc1r1
|
host cell factor C1 regulator 1 |
| chr11_+_32211115 | 0.06 |
ENSRNOT00000087452
|
Mrps6
|
mitochondrial ribosomal protein S6 |
| chr13_+_75111778 | 0.06 |
ENSRNOT00000006924
|
Tp53i3
|
tumor protein p53 inducible protein 3 |
| chr3_+_151285249 | 0.06 |
ENSRNOT00000055254
|
Procr
|
protein C receptor |
| chr15_+_50891127 | 0.06 |
ENSRNOT00000020728
|
Stc1
|
stanniocalcin 1 |
| chr1_+_196942364 | 0.06 |
ENSRNOT00000088073
ENSRNOT00000020994 |
Il4r
|
interleukin 4 receptor |
| chr10_+_85744568 | 0.06 |
ENSRNOT00000005522
|
Lasp1
|
LIM and SH3 protein 1 |
| chr13_+_85818427 | 0.06 |
ENSRNOT00000077227
ENSRNOT00000006117 |
Rxrg
|
retinoid X receptor gamma |
| chr16_-_36161089 | 0.06 |
ENSRNOT00000017888
|
Scrg1
|
stimulator of chondrogenesis 1 |
| chr3_+_152752091 | 0.06 |
ENSRNOT00000037177
|
Dlgap4
|
DLG associated protein 4 |
| chr13_-_50499060 | 0.06 |
ENSRNOT00000065347
ENSRNOT00000076924 |
Etnk2
|
ethanolamine kinase 2 |
| chr1_-_142759882 | 0.06 |
ENSRNOT00000015218
|
Sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr7_+_11077411 | 0.06 |
ENSRNOT00000007117
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr10_-_108196217 | 0.06 |
ENSRNOT00000075440
|
Cbx4
|
chromobox 4 |
| chr5_-_58113553 | 0.06 |
ENSRNOT00000046589
|
Arid3c
|
AT-rich interaction domain 3C |
| chr5_-_155916893 | 0.06 |
ENSRNOT00000055947
|
Ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr14_+_86922223 | 0.06 |
ENSRNOT00000086468
|
Ramp3
|
receptor activity modifying protein 3 |
| chrX_-_68562301 | 0.06 |
ENSRNOT00000076720
|
Ophn1
|
oligophrenin 1 |
| chr11_-_61530567 | 0.06 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr5_-_153924896 | 0.06 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.3 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.2 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.2 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.1 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.0 | 0.1 | GO:1904638 | response to resveratrol(GO:1904638) |
| 0.0 | 0.2 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.2 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.0 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.0 | 0.0 | GO:0097214 | regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) |
| 0.0 | 0.1 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:0090194 | negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.1 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.0 | 0.1 | GO:0014736 | negative regulation of muscle atrophy(GO:0014736) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0032639 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.2 | GO:0048597 | B cell selection(GO:0002339) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.2 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0034756 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.0 | 0.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.0 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.1 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.0 | 0.1 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 0.0 | 0.0 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.5 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.1 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.0 | 0.0 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:2000853 | negative regulation of corticosterone secretion(GO:2000853) |
| 0.0 | 0.1 | GO:1903002 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.1 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.0 | 0.0 | GO:0032805 | positive regulation of low-density lipoprotein particle receptor catabolic process(GO:0032805) |
| 0.0 | 0.1 | GO:0001810 | regulation of type I hypersensitivity(GO:0001810) type I hypersensitivity(GO:0016068) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.1 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.0 | GO:0060468 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) |
| 0.0 | 0.0 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:1904170 | regulation of bleb assembly(GO:1904170) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.1 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0031659 | positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.0 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.0 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.0 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.0 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0051586 | glial cell-derived neurotrophic factor secretion(GO:0044467) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) negative regulation of dopamine receptor signaling pathway(GO:0060160) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.0 | 0.0 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 0.0 | 0.0 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.1 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.0 | 0.0 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.0 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.0 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.0 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.0 | GO:0015746 | citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.0 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.0 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:0021892 | cerebral cortex GABAergic interneuron differentiation(GO:0021892) |
| 0.0 | 0.0 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.0 | 0.1 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.0 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.0 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.2 | GO:0036398 | TCR signalosome(GO:0036398) microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.0 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.0 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.0 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.0 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 1.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0030613 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 0.0 | 0.0 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.0 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.0 | GO:0047619 | acylcarnitine hydrolase activity(GO:0047619) |
| 0.0 | 0.0 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.0 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.0 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.0 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |