Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
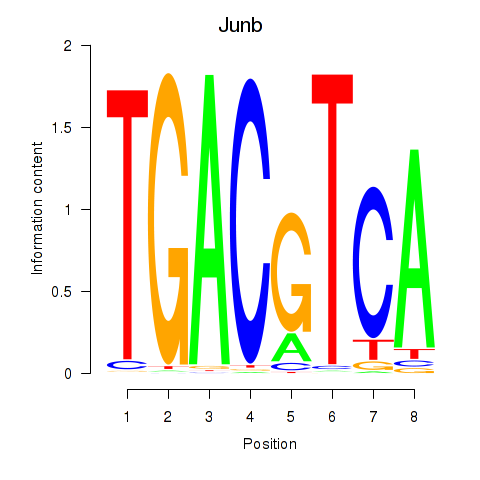
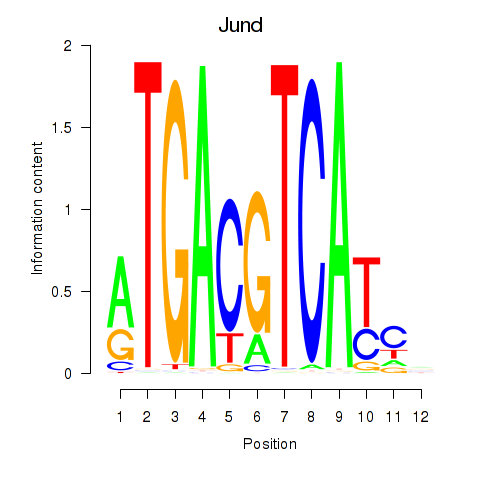
Results for Junb_Jund
Z-value: 1.16
Transcription factors associated with Junb_Jund
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Junb
|
ENSRNOG00000042838 | JunB proto-oncogene, AP-1 transcription factor subunit |
|
Jund
|
ENSRNOG00000019568 | JunD proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Jund | rn6_v1_chr16_-_20486707_20486707 | 0.90 | 4.0e-02 | Click! |
| Junb | rn6_v1_chr19_-_26094756_26094756 | 0.83 | 8.0e-02 | Click! |
Activity profile of Junb_Jund motif
Sorted Z-values of Junb_Jund motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_9695292 | 3.36 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr7_+_120580743 | 1.48 |
ENSRNOT00000017181
|
Maff
|
MAF bZIP transcription factor F |
| chr7_-_44121130 | 0.74 |
ENSRNOT00000005706
|
Nts
|
neurotensin |
| chr10_+_16970626 | 0.68 |
ENSRNOT00000005383
|
Dusp1
|
dual specificity phosphatase 1 |
| chr16_-_19399851 | 0.65 |
ENSRNOT00000089056
ENSRNOT00000021073 |
Tpm4
|
tropomyosin 4 |
| chr10_-_85574889 | 0.65 |
ENSRNOT00000072274
|
LOC691153
|
hypothetical protein LOC691153 |
| chr6_+_146784915 | 0.60 |
ENSRNOT00000008362
|
Sp8
|
Sp8 transcription factor |
| chr17_+_57074525 | 0.58 |
ENSRNOT00000020012
ENSRNOT00000074146 |
Crem
|
cAMP responsive element modulator |
| chr5_-_152464850 | 0.58 |
ENSRNOT00000021937
|
Zfp593
|
zinc finger protein 593 |
| chr12_-_13668515 | 0.56 |
ENSRNOT00000086847
|
Fscn1
|
fascin actin-bundling protein 1 |
| chr2_+_164549455 | 0.55 |
ENSRNOT00000017151
|
Mlf1
|
myeloid leukemia factor 1 |
| chr8_+_114866768 | 0.52 |
ENSRNOT00000076731
|
Wdr82
|
WD repeat domain 82 |
| chr1_-_216080287 | 0.52 |
ENSRNOT00000027682
|
Th
|
tyrosine hydroxylase |
| chr16_-_20486707 | 0.51 |
ENSRNOT00000026470
|
Jund
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr4_+_83391283 | 0.50 |
ENSRNOT00000031365
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr17_+_57075218 | 0.47 |
ENSRNOT00000089536
|
Crem
|
cAMP responsive element modulator |
| chr9_+_102862890 | 0.47 |
ENSRNOT00000050494
ENSRNOT00000080129 |
Fam174a
|
family with sequence similarity 174, member A |
| chr11_+_31389514 | 0.46 |
ENSRNOT00000000325
|
Olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr4_+_8256611 | 0.46 |
ENSRNOT00000061894
|
AABR07059198.1
|
|
| chr7_+_123168811 | 0.45 |
ENSRNOT00000007091
|
Csdc2
|
cold shock domain containing C2 |
| chr5_+_50381244 | 0.42 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr1_+_224938801 | 0.40 |
ENSRNOT00000073052
|
1700092M07Rik
|
RIKEN cDNA 1700092M07 gene |
| chr17_-_43821536 | 0.39 |
ENSRNOT00000072286
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr1_+_73719005 | 0.39 |
ENSRNOT00000025110
|
Cdc42ep5
|
CDC42 effector protein 5 |
| chr2_-_30127269 | 0.39 |
ENSRNOT00000023869
|
Cartpt
|
CART prepropeptide |
| chr9_-_78969013 | 0.37 |
ENSRNOT00000019772
ENSRNOT00000057585 |
Fn1
|
fibronectin 1 |
| chr9_-_85243001 | 0.37 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr16_+_72388880 | 0.37 |
ENSRNOT00000072459
|
LOC684871
|
similar to Protein C8orf4 (Thyroid cancer protein 1) (TC-1) |
| chr6_+_98284170 | 0.36 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr2_-_104461863 | 0.36 |
ENSRNOT00000016953
|
Crh
|
corticotropin releasing hormone |
| chr13_-_50499060 | 0.35 |
ENSRNOT00000065347
ENSRNOT00000076924 |
Etnk2
|
ethanolamine kinase 2 |
| chr16_+_72401887 | 0.35 |
ENSRNOT00000074449
|
LOC100910163
|
uncharacterized LOC100910163 |
| chr11_-_36479868 | 0.34 |
ENSRNOT00000075762
|
LOC100911295
|
non-histone chromosomal protein HMG-14-like |
| chr10_-_85435016 | 0.34 |
ENSRNOT00000079921
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr11_+_88122271 | 0.33 |
ENSRNOT00000002540
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr3_+_150801289 | 0.32 |
ENSRNOT00000035060
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr20_+_5080070 | 0.32 |
ENSRNOT00000086950
ENSRNOT00000077082 |
Abhd16a
|
abhydrolase domain containing 16A |
| chrX_-_45522665 | 0.32 |
ENSRNOT00000030771
|
RGD1562200
|
similar to GS2 gene |
| chr12_-_18531161 | 0.31 |
ENSRNOT00000037829
|
Akap17a
|
A-kinase anchoring protein 17A |
| chr19_+_56272162 | 0.31 |
ENSRNOT00000030399
|
Afg3l1
|
AFG3(ATPase family gene 3)-like 1 (S. cerevisiae) |
| chr5_+_159845774 | 0.31 |
ENSRNOT00000012328
|
Epha2
|
Eph receptor A2 |
| chr20_+_4966817 | 0.30 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr12_+_52637000 | 0.30 |
ENSRNOT00000063905
|
Gtpbp6
|
GTP binding protein 6 (putative) |
| chr6_+_1657331 | 0.28 |
ENSRNOT00000049672
ENSRNOT00000079864 |
Qpct
|
glutaminyl-peptide cyclotransferase |
| chr17_+_57040023 | 0.26 |
ENSRNOT00000020204
|
Crem
|
cAMP responsive element modulator |
| chr7_-_58587787 | 0.25 |
ENSRNOT00000005814
|
Lgr5
|
leucine rich repeat containing G protein coupled receptor 5 |
| chr3_+_3767394 | 0.25 |
ENSRNOT00000067840
|
Gpsm1
|
G-protein signaling modulator 1 |
| chr5_-_151397603 | 0.24 |
ENSRNOT00000076866
|
Gpr3
|
G protein-coupled receptor 3 |
| chr1_+_86938138 | 0.24 |
ENSRNOT00000075601
|
Ccer2
|
coiled-coil glutamate-rich protein 2 |
| chr4_+_168832910 | 0.24 |
ENSRNOT00000011134
|
Gprc5a
|
G protein-coupled receptor, class C, group 5, member A |
| chr5_-_114014277 | 0.24 |
ENSRNOT00000011731
|
Jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr17_+_31493107 | 0.24 |
ENSRNOT00000023611
ENSRNOT00000086264 |
Tubb2a
|
tubulin, beta 2A class IIa |
| chr2_+_124400679 | 0.24 |
ENSRNOT00000058495
ENSRNOT00000067034 |
Spry1
|
sprouty RTK signaling antagonist 1 |
| chr7_+_72924799 | 0.23 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr6_+_26602144 | 0.23 |
ENSRNOT00000008037
|
Ucn
|
urocortin |
| chr12_+_52670898 | 0.23 |
ENSRNOT00000056632
|
LOC100912590
|
putative GTP-binding protein 6-like |
| chr7_-_2909144 | 0.23 |
ENSRNOT00000082518
ENSRNOT00000089074 ENSRNOT00000085644 |
Myl6
|
myosin light chain 6 |
| chr13_+_52553775 | 0.23 |
ENSRNOT00000011991
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr2_+_188784222 | 0.22 |
ENSRNOT00000028095
|
Pmvk
|
phosphomevalonate kinase |
| chr14_-_6813945 | 0.22 |
ENSRNOT00000002938
|
Ibsp
|
integrin-binding sialoprotein |
| chr16_-_49574314 | 0.22 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr1_-_47502952 | 0.22 |
ENSRNOT00000025580
|
Tagap
|
T-cell activation RhoGTPase activating protein |
| chr3_+_176093640 | 0.22 |
ENSRNOT00000086541
|
Ogfr
|
opioid growth factor receptor |
| chr4_+_99239115 | 0.22 |
ENSRNOT00000009515
|
Cd8a
|
CD8a molecule |
| chr9_+_82556573 | 0.22 |
ENSRNOT00000026860
|
Des
|
desmin |
| chr3_+_175930505 | 0.22 |
ENSRNOT00000012611
|
Ogfr
|
opioid growth factor receptor |
| chr5_-_153625869 | 0.22 |
ENSRNOT00000024464
|
Clic4
|
chloride intracellular channel 4 |
| chr9_+_10535340 | 0.21 |
ENSRNOT00000075408
|
Znrf4
|
zinc and ring finger 4 |
| chr2_+_187347602 | 0.21 |
ENSRNOT00000025384
|
Nes
|
nestin |
| chr1_-_80221710 | 0.21 |
ENSRNOT00000091687
|
Fosb
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr8_+_48472824 | 0.21 |
ENSRNOT00000010463
ENSRNOT00000090780 |
Mcam
|
melanoma cell adhesion molecule |
| chr5_+_58393603 | 0.21 |
ENSRNOT00000080082
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr1_-_188190778 | 0.21 |
ENSRNOT00000092657
ENSRNOT00000022988 |
Coq7
|
coenzyme Q7, hydroxylase |
| chr10_-_13542077 | 0.20 |
ENSRNOT00000008736
|
Atp6v0c
|
ATPase H+ transporting V0 subunit C |
| chr20_-_10680283 | 0.20 |
ENSRNOT00000001579
|
Sik1
|
salt-inducible kinase 1 |
| chr11_+_88424414 | 0.20 |
ENSRNOT00000022328
|
Spag6l
|
sperm associated antigen 6-like |
| chr8_+_64364741 | 0.20 |
ENSRNOT00000082840
|
Celf6
|
CUGBP, Elav-like family member 6 |
| chr13_+_104284660 | 0.20 |
ENSRNOT00000005400
|
Dusp10
|
dual specificity phosphatase 10 |
| chr15_-_6587367 | 0.20 |
ENSRNOT00000038449
|
Zfp385d
|
zinc finger protein 385D |
| chr16_+_20317446 | 0.20 |
ENSRNOT00000025626
|
Ccdc124
|
coiled-coil domain containing 124 |
| chr1_-_89303968 | 0.19 |
ENSRNOT00000056714
|
Ffar3
|
free fatty acid receptor 3 |
| chr1_-_209641123 | 0.19 |
ENSRNOT00000021702
|
Ebf3
|
early B-cell factor 3 |
| chr1_-_64090017 | 0.19 |
ENSRNOT00000086622
ENSRNOT00000091654 |
Rps9
|
ribosomal protein S9 |
| chr12_-_2007516 | 0.19 |
ENSRNOT00000037564
|
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr19_+_28965583 | 0.19 |
ENSRNOT00000049865
|
LOC685226
|
similar to spermatogenesis associated glutamate (E)-rich protein 4d |
| chr20_+_13732198 | 0.19 |
ENSRNOT00000008608
|
LOC103694877
|
macrophage migration inhibitory factor |
| chr2_-_104133985 | 0.19 |
ENSRNOT00000088167
ENSRNOT00000067725 |
Pde7a
|
phosphodiesterase 7A |
| chr9_+_82674202 | 0.18 |
ENSRNOT00000027208
|
Tmem198
|
transmembrane protein 198 |
| chr15_+_43007908 | 0.18 |
ENSRNOT00000084753
ENSRNOT00000091567 ENSRNOT00000087709 |
Stmn4
|
stathmin 4 |
| chr5_+_58393233 | 0.18 |
ENSRNOT00000000142
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr9_-_82673898 | 0.18 |
ENSRNOT00000027165
|
Chpf
|
chondroitin polymerizing factor |
| chr14_-_6900733 | 0.18 |
ENSRNOT00000061224
|
Dmp1
|
dentin matrix acidic phosphoprotein 1 |
| chr1_+_201429771 | 0.18 |
ENSRNOT00000027836
|
Plekha1
|
pleckstrin homology domain containing A1 |
| chr1_-_101514974 | 0.18 |
ENSRNOT00000044788
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr20_+_29655226 | 0.17 |
ENSRNOT00000089059
|
Spock2
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 2 |
| chr3_-_7498555 | 0.17 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr1_-_220467159 | 0.17 |
ENSRNOT00000075365
|
Tmem151a
|
transmembrane protein 151A |
| chr10_-_66020682 | 0.17 |
ENSRNOT00000011019
|
Fam58b
|
family with sequence similarity 58, member B |
| chr10_-_29450644 | 0.17 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr6_-_103313074 | 0.16 |
ENSRNOT00000083677
|
Zfp36l1
|
zinc finger protein 36, C3H type-like 1 |
| chr3_-_160922341 | 0.16 |
ENSRNOT00000029206
|
Tp53tg5
|
TP53 target 5 |
| chr12_-_18530724 | 0.16 |
ENSRNOT00000078999
|
Akap17a
|
A-kinase anchoring protein 17A |
| chr6_-_46631983 | 0.16 |
ENSRNOT00000045963
|
Sox11
|
SRY box 11 |
| chr7_+_121311024 | 0.16 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr6_-_21600451 | 0.16 |
ENSRNOT00000047674
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr15_+_52265557 | 0.15 |
ENSRNOT00000015969
|
Nudt18
|
nudix hydrolase 18 |
| chr14_-_18531179 | 0.15 |
ENSRNOT00000003703
|
Areg
|
amphiregulin |
| chr3_+_37545238 | 0.15 |
ENSRNOT00000070792
|
Tnfaip6
|
TNF alpha induced protein 6 |
| chr10_+_70884531 | 0.15 |
ENSRNOT00000015199
|
Ccl4
|
C-C motif chemokine ligand 4 |
| chr1_+_166428761 | 0.15 |
ENSRNOT00000085555
|
Stard10
|
StAR-related lipid transfer domain containing 10 |
| chr4_+_27175243 | 0.15 |
ENSRNOT00000009985
|
Cyp51
|
cytochrome P450, family 51 |
| chr6_+_126434226 | 0.15 |
ENSRNOT00000090857
|
Chga
|
chromogranin A |
| chr4_-_50312608 | 0.15 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr2_+_233602732 | 0.14 |
ENSRNOT00000044232
|
Pitx2
|
paired-like homeodomain 2 |
| chr19_+_55917736 | 0.14 |
ENSRNOT00000020635
|
Rpl13
|
ribosomal protein L13 |
| chr1_-_198382614 | 0.14 |
ENSRNOT00000055016
|
Asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr14_+_108826831 | 0.14 |
ENSRNOT00000083146
ENSRNOT00000009421 |
Bcl11a
|
B-cell CLL/lymphoma 11A |
| chr6_+_132702448 | 0.14 |
ENSRNOT00000005743
|
Yy1
|
YY1 transcription factor |
| chr11_+_39482408 | 0.14 |
ENSRNOT00000075126
|
Hmgn1
|
high mobility group nucleosome binding domain 1 |
| chr1_-_64021321 | 0.14 |
ENSRNOT00000090819
|
Rps9
|
ribosomal protein S9 |
| chr1_+_201913148 | 0.14 |
ENSRNOT00000036128
|
Fam24a
|
family with sequence similarity 24, member A |
| chr1_-_89179825 | 0.14 |
ENSRNOT00000028514
|
Tmem147
|
transmembrane protein 147 |
| chr8_+_85489553 | 0.13 |
ENSRNOT00000082158
|
Rpl12-ps1
|
ribosomal protein L12, pseudogene 1 |
| chr3_-_120011364 | 0.13 |
ENSRNOT00000018922
|
Fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr1_-_89194602 | 0.13 |
ENSRNOT00000028518
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr1_+_221735517 | 0.13 |
ENSRNOT00000028628
ENSRNOT00000044866 |
Sf1
|
splicing factor 1 |
| chr17_+_10537365 | 0.13 |
ENSRNOT00000023651
|
Cltb
|
clathrin, light chain B |
| chr11_-_33003021 | 0.13 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr5_+_147692391 | 0.13 |
ENSRNOT00000064855
|
Fam229a
|
family with sequence similarity 229, member A |
| chr2_+_86951776 | 0.13 |
ENSRNOT00000087275
|
AABR07009105.1
|
|
| chr19_+_27404712 | 0.13 |
ENSRNOT00000023657
|
Mylk3
|
myosin light chain kinase 3 |
| chr14_-_108284619 | 0.13 |
ENSRNOT00000078814
|
LOC690352
|
hypothetical protein LOC690352 |
| chr5_-_146446227 | 0.13 |
ENSRNOT00000044868
|
Hmgb4
|
high-mobility group box 4 |
| chr14_-_86739335 | 0.12 |
ENSRNOT00000078282
|
Purb
|
purine rich element binding protein B |
| chr19_+_27835890 | 0.12 |
ENSRNOT00000042150
|
LOC100909409
|
disks large homolog 5-like |
| chr4_-_184096806 | 0.12 |
ENSRNOT00000055433
|
LOC100362344
|
mKIAA1238 protein-like |
| chr3_+_163552166 | 0.12 |
ENSRNOT00000049082
|
Tp53rk
|
TP53 regulating kinase |
| chr15_-_70399924 | 0.12 |
ENSRNOT00000087940
|
Diaph3
|
diaphanous-related formin 3 |
| chr9_+_17817721 | 0.12 |
ENSRNOT00000086986
ENSRNOT00000026920 |
Hsp90ab1
|
heat shock protein 90 alpha family class B member 1 |
| chr1_-_80544825 | 0.12 |
ENSRNOT00000057802
ENSRNOT00000040060 ENSRNOT00000067049 ENSRNOT00000052387 ENSRNOT00000073352 |
Relb
|
RELB proto-oncogene, NF-kB subunit |
| chr1_+_100473643 | 0.12 |
ENSRNOT00000026379
|
Josd2
|
Josephin domain containing 2 |
| chr3_+_150910398 | 0.12 |
ENSRNOT00000055310
|
Tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr3_+_79918969 | 0.12 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr1_-_101514547 | 0.12 |
ENSRNOT00000079633
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr1_+_191344979 | 0.12 |
ENSRNOT00000023773
|
Hs3st2
|
heparan sulfate-glucosamine 3-sulfotransferase 2 |
| chr4_+_157453069 | 0.12 |
ENSRNOT00000088622
|
Mlf2
|
myeloid leukemia factor 2 |
| chr13_+_49003803 | 0.12 |
ENSRNOT00000080556
|
Lemd1
|
LEM domain containing 1 |
| chr10_-_57064600 | 0.12 |
ENSRNOT00000032926
|
Cxcl16
|
C-X-C motif chemokine ligand 16 |
| chr4_+_157452607 | 0.12 |
ENSRNOT00000022467
|
Mlf2
|
myeloid leukemia factor 2 |
| chr4_-_181477281 | 0.12 |
ENSRNOT00000055463
|
Mansc4
|
MANSC domain containing 4 |
| chr20_-_3299420 | 0.12 |
ENSRNOT00000090999
|
Gnl1
|
G protein nucleolar 1 |
| chr1_-_52495582 | 0.12 |
ENSRNOT00000067142
|
Pde10a
|
phosphodiesterase 10A |
| chr1_+_84304228 | 0.11 |
ENSRNOT00000024771
|
Prx
|
periaxin |
| chr3_+_125428260 | 0.11 |
ENSRNOT00000028892
|
Chgb
|
chromogranin B |
| chr9_-_16406338 | 0.11 |
ENSRNOT00000073079
|
Tbcc
|
tubulin folding cofactor C |
| chr7_+_141054494 | 0.11 |
ENSRNOT00000088176
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr12_+_2170630 | 0.11 |
ENSRNOT00000071928
|
Pet100
|
PET100 homolog |
| chrX_-_139329975 | 0.11 |
ENSRNOT00000086405
|
LOC103694517
|
high mobility group protein B4-like |
| chr19_-_36157924 | 0.11 |
ENSRNOT00000072022
|
AABR07043701.1
|
|
| chr8_+_48094673 | 0.11 |
ENSRNOT00000008614
|
Nectin1
|
nectin cell adhesion molecule 1 |
| chr4_-_120559078 | 0.11 |
ENSRNOT00000085730
ENSRNOT00000079575 |
Kbtbd12
|
kelch repeat and BTB domain containing 12 |
| chr19_-_56731372 | 0.11 |
ENSRNOT00000024182
|
Nup133
|
nucleoporin 133 |
| chr2_+_233615739 | 0.11 |
ENSRNOT00000051009
|
Pitx2
|
paired-like homeodomain 2 |
| chr6_+_136279496 | 0.11 |
ENSRNOT00000091946
|
Apopt1
|
apoptogenic 1, mitochondrial |
| chr6_+_136185487 | 0.10 |
ENSRNOT00000015373
|
Apopt1
|
apoptogenic 1, mitochondrial |
| chr8_+_23113048 | 0.10 |
ENSRNOT00000029577
|
Cnn1
|
calponin 1 |
| chr10_-_102269272 | 0.10 |
ENSRNOT00000048264
|
Cdc42ep4
|
CDC42 effector protein 4 |
| chr14_+_107767392 | 0.10 |
ENSRNOT00000012847
|
Cct4
|
chaperonin containing TCP1 subunit 4 |
| chr18_-_14016713 | 0.10 |
ENSRNOT00000041125
|
Nol4
|
nucleolar protein 4 |
| chr14_-_86868598 | 0.10 |
ENSRNOT00000087212
|
Nacad
|
NAC alpha domain containing |
| chr1_-_81295442 | 0.10 |
ENSRNOT00000030900
|
Irgc
|
immunity related GTPase cinema |
| chr8_+_22021213 | 0.10 |
ENSRNOT00000049706
|
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr20_-_3299580 | 0.10 |
ENSRNOT00000050373
|
Gnl1
|
G protein nucleolar 1 |
| chr1_-_100501437 | 0.10 |
ENSRNOT00000026445
|
Emc10
|
ER membrane protein complex subunit 10 |
| chr5_+_78428669 | 0.10 |
ENSRNOT00000080744
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr10_+_89376530 | 0.10 |
ENSRNOT00000028089
|
Rnd2
|
Rho family GTPase 2 |
| chr14_-_83741969 | 0.10 |
ENSRNOT00000026293
|
Inpp5j
|
inositol polyphosphate-5-phosphatase J |
| chr4_-_157252565 | 0.09 |
ENSRNOT00000079947
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chrX_-_1346181 | 0.09 |
ENSRNOT00000042736
|
LOC102555453
|
60S ribosomal protein L12-like |
| chr2_+_45104305 | 0.09 |
ENSRNOT00000014559
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr12_-_18526250 | 0.09 |
ENSRNOT00000001791
|
Asmt
|
acetylserotonin O-methyltransferase |
| chr1_+_100501676 | 0.09 |
ENSRNOT00000043724
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr19_-_54245855 | 0.09 |
ENSRNOT00000023855
|
Emc8
|
ER membrane protein complex subunit 8 |
| chr4_+_84854386 | 0.09 |
ENSRNOT00000013620
|
Mturn
|
maturin, neural progenitor differentiation regulator homolog |
| chr10_+_89578212 | 0.09 |
ENSRNOT00000028178
|
Arl4d
|
ADP-ribosylation factor like GTPase 4D |
| chr17_+_10463303 | 0.09 |
ENSRNOT00000060822
|
Rnf44
|
ring finger protein 44 |
| chr7_-_18612118 | 0.09 |
ENSRNOT00000078122
ENSRNOT00000010197 |
Rab11b
|
RAB11B, member RAS oncogene family |
| chr7_-_12085226 | 0.09 |
ENSRNOT00000088669
ENSRNOT00000079925 |
Onecut3
|
one cut domain, family member 3 |
| chr12_+_37984790 | 0.09 |
ENSRNOT00000001445
|
Vps37b
|
VPS37B, ESCRT-I subunit |
| chr2_-_66608324 | 0.09 |
ENSRNOT00000077597
|
Cln5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr7_-_120518653 | 0.09 |
ENSRNOT00000016362
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr10_+_37594824 | 0.09 |
ENSRNOT00000085463
|
Skp1
|
S-phase kinase-associated protein 1 |
| chr20_+_3299730 | 0.09 |
ENSRNOT00000078538
|
Prr3
|
proline rich 3 |
| chr1_+_78015722 | 0.09 |
ENSRNOT00000002043
|
Kptn
|
kaptin (actin binding protein) |
| chr6_+_72891725 | 0.09 |
ENSRNOT00000038074
|
Nubpl
|
nucleotide binding protein-like |
| chr7_+_10962330 | 0.09 |
ENSRNOT00000008360
|
Tle6
|
transducin-like enhancer of split 6 |
| chr1_-_198267093 | 0.09 |
ENSRNOT00000047477
|
RGD1563217
|
similar to RIKEN cDNA 4930451I11 |
| chr6_+_26387877 | 0.09 |
ENSRNOT00000076105
|
Fndc4
|
|
| chr16_-_20873344 | 0.08 |
ENSRNOT00000027381
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr2_+_189423559 | 0.08 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Junb_Jund
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.2 | 0.5 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.7 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.1 | 0.5 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.4 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.1 | 0.3 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.3 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) response to environmental enrichment(GO:0090648) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.6 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.3 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.1 | 0.3 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.3 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.2 | GO:0060935 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.1 | 0.6 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.2 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.1 | 0.2 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.1 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.1 | 0.2 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 0.2 | GO:0045819 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 | 0.2 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) mesendoderm development(GO:0048382) |
| 0.1 | 0.2 | GO:0045105 | intermediate filament polymerization or depolymerization(GO:0045105) |
| 0.1 | 0.2 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.1 | 0.2 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.1 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.4 | GO:1990441 | negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990441) |
| 0.0 | 0.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.3 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.7 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.0 | 0.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.6 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 0.0 | 1.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.5 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 1.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.0 | 0.2 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 | 0.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.3 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.0 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.5 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:1904685 | positive regulation of skeletal muscle cell proliferation(GO:0014858) positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.0 | 0.2 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.2 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.1 | GO:1903576 | response to L-arginine(GO:1903576) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.0 | GO:0030167 | proteoglycan catabolic process(GO:0030167) heparan sulfate proteoglycan catabolic process(GO:0030200) positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.1 | GO:1903659 | complement-dependent cytotoxicity(GO:0097278) regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.1 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.1 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.0 | 0.0 | GO:0009216 | purine deoxyribonucleotide biosynthetic process(GO:0009153) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.0 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.0 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.0 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.1 | GO:0046070 | dGTP metabolic process(GO:0046070) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.0 | GO:1902869 | regulation of amacrine cell differentiation(GO:1902869) |
| 0.0 | 0.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.6 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 0.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0045252 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0099571 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.0 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.0 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 0.6 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.6 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.3 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.3 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.4 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 0.2 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 1.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:0017098 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0052743 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.0 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.0 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |