Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Isl2
Z-value: 0.72
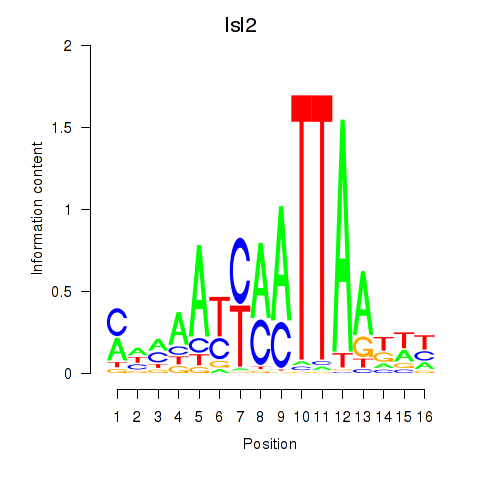
Transcription factors associated with Isl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Isl2
|
ENSRNOG00000015336 | ISL LIM homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isl2 | rn6_v1_chr8_+_60117729_60117729 | 0.44 | 4.5e-01 | Click! |
Activity profile of Isl2 motif
Sorted Z-values of Isl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_91054974 | 0.23 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr7_+_144503534 | 0.23 |
ENSRNOT00000021543
|
Tbca
|
tubulin folding cofactor A |
| chr13_-_83457888 | 0.22 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr7_+_2300434 | 0.21 |
ENSRNOT00000071785
|
AABR07055413.1
|
|
| chr8_+_130366775 | 0.20 |
ENSRNOT00000071152
|
Nktr
|
natural killer cell triggering receptor |
| chr11_+_15081774 | 0.19 |
ENSRNOT00000009911
|
LOC108348050
|
ubiquitin carboxyl-terminal hydrolase 25-like |
| chr1_-_252808380 | 0.18 |
ENSRNOT00000025856
|
Ch25h
|
cholesterol 25-hydroxylase |
| chr9_+_73378057 | 0.18 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr4_+_61814974 | 0.16 |
ENSRNOT00000074951
|
Akr1b10
|
aldo-keto reductase family 1 member B10 |
| chr2_+_86996497 | 0.16 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr1_+_192379543 | 0.16 |
ENSRNOT00000078705
|
Prkcb
|
protein kinase C, beta |
| chr3_+_63379031 | 0.16 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr4_-_155690869 | 0.15 |
ENSRNOT00000012216
|
C3ar1
|
complement C3a receptor 1 |
| chr1_+_145770135 | 0.15 |
ENSRNOT00000015320
|
Stard5
|
StAR-related lipid transfer domain containing 5 |
| chr1_-_78180216 | 0.14 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr2_+_23289374 | 0.14 |
ENSRNOT00000090666
ENSRNOT00000032783 |
Dmgdh
|
dimethylglycine dehydrogenase |
| chr3_-_60765645 | 0.14 |
ENSRNOT00000050513
|
Atf2
|
activating transcription factor 2 |
| chr18_-_75207306 | 0.14 |
ENSRNOT00000021717
|
Setbp1
|
SET binding protein 1 |
| chr18_+_1723565 | 0.14 |
ENSRNOT00000033468
|
Greb1l
|
growth regulation by estrogen in breast cancer 1 like |
| chr14_+_17228856 | 0.14 |
ENSRNOT00000003082
|
Cxcl9
|
C-X-C motif chemokine ligand 9 |
| chr15_+_87722221 | 0.14 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr2_-_27343998 | 0.13 |
ENSRNOT00000091418
|
Polk
|
DNA polymerase kappa |
| chr16_+_84465656 | 0.13 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr18_+_74299931 | 0.13 |
ENSRNOT00000078403
|
Epg5
|
ectopic P-granules autophagy protein 5 homolog |
| chr9_+_66889028 | 0.13 |
ENSRNOT00000087194
|
Carf
|
calcium responsive transcription factor |
| chr14_-_77810147 | 0.13 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chr3_-_165700489 | 0.12 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr3_+_113918629 | 0.12 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr1_-_38538987 | 0.12 |
ENSRNOT00000090406
ENSRNOT00000071275 |
LOC102551340
|
zinc finger protein 728-like |
| chr2_+_86996798 | 0.12 |
ENSRNOT00000070821
|
Zfp455
|
zinc finger protein 455 |
| chr3_+_48626038 | 0.11 |
ENSRNOT00000009697
|
Gca
|
grancalcin |
| chr12_+_25435567 | 0.11 |
ENSRNOT00000092982
|
Gtf2i
|
general transcription factor II I |
| chr12_-_35979193 | 0.11 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr9_+_73418607 | 0.11 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr1_+_116604550 | 0.11 |
ENSRNOT00000083587
|
Ube3a
|
ubiquitin protein ligase E3A |
| chr3_-_138683318 | 0.11 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr9_+_61823531 | 0.11 |
ENSRNOT00000070844
|
Mars2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr6_-_111477090 | 0.11 |
ENSRNOT00000016389
|
Alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr7_+_24638208 | 0.10 |
ENSRNOT00000009135
|
Mterf2
|
mitochondrial transcription termination factor 2 |
| chr1_+_253221812 | 0.10 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chr1_+_101603222 | 0.10 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr1_-_43638161 | 0.10 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr3_+_48096954 | 0.10 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr10_-_36323055 | 0.10 |
ENSRNOT00000081026
|
Zfp354c
|
zinc finger protein 354C |
| chr4_+_66090209 | 0.10 |
ENSRNOT00000031193
|
Ttc26
|
tetratricopeptide repeat domain 26 |
| chr9_-_80166807 | 0.10 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr5_-_19559244 | 0.10 |
ENSRNOT00000014289
ENSRNOT00000089666 |
Nsmaf
|
neutral sphingomyelinase activation associated factor |
| chr3_+_131351587 | 0.10 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr1_+_61786900 | 0.09 |
ENSRNOT00000090287
|
AABR07001905.1
|
|
| chr4_+_88694583 | 0.09 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr13_+_98311827 | 0.09 |
ENSRNOT00000082844
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chrX_+_14019961 | 0.09 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr16_+_10417185 | 0.09 |
ENSRNOT00000082186
|
Anxa8
|
annexin A8 |
| chr7_+_42304534 | 0.09 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr15_-_55209342 | 0.09 |
ENSRNOT00000021752
|
Rb1
|
RB transcriptional corepressor 1 |
| chr7_-_139649286 | 0.09 |
ENSRNOT00000080056
|
Senp1
|
SUMO1/sentrin specific peptidase 1 |
| chr7_+_15422479 | 0.09 |
ENSRNOT00000066520
|
Zfp563
|
zinc finger protein 563 |
| chr1_+_252409268 | 0.09 |
ENSRNOT00000026219
|
Lipm
|
lipase, family member M |
| chr15_-_88670349 | 0.09 |
ENSRNOT00000012881
|
Rnf219
|
ring finger protein 219 |
| chr6_-_42002819 | 0.09 |
ENSRNOT00000032417
|
Greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr10_+_84966989 | 0.09 |
ENSRNOT00000013580
|
Scrn2
|
secernin 2 |
| chr1_+_80028928 | 0.09 |
ENSRNOT00000012082
|
Fbxo46
|
F-box protein 46 |
| chr8_-_21519013 | 0.09 |
ENSRNOT00000042903
|
Zfp266
|
zinc finger protein 266 |
| chr2_+_209661244 | 0.09 |
ENSRNOT00000091973
|
AABR07012826.1
|
|
| chr4_+_31229913 | 0.09 |
ENSRNOT00000077134
ENSRNOT00000087897 |
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr6_-_108329464 | 0.08 |
ENSRNOT00000016040
|
Abcd4
|
ATP binding cassette subfamily D member 4 |
| chr8_+_133197032 | 0.08 |
ENSRNOT00000075564
|
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr13_+_34365147 | 0.08 |
ENSRNOT00000093066
|
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr17_+_45467015 | 0.08 |
ENSRNOT00000081408
|
Gpx5
|
glutathione peroxidase 5 |
| chr17_+_53965562 | 0.08 |
ENSRNOT00000090387
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr12_-_1195591 | 0.08 |
ENSRNOT00000001446
|
Stard13
|
StAR-related lipid transfer domain containing 13 |
| chr3_-_66885085 | 0.08 |
ENSRNOT00000084299
ENSRNOT00000090547 |
Pde1a
|
phosphodiesterase 1A |
| chr2_+_2729829 | 0.08 |
ENSRNOT00000017068
|
Spata9
|
spermatogenesis associated 9 |
| chr6_+_18880737 | 0.08 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr10_-_10881844 | 0.08 |
ENSRNOT00000087118
ENSRNOT00000004416 |
Mgrn1
|
mahogunin ring finger 1 |
| chr13_+_105684420 | 0.08 |
ENSRNOT00000040543
|
Gpatch2
|
G patch domain containing 2 |
| chr17_-_56068125 | 0.08 |
ENSRNOT00000022462
|
Mtpap
|
mitochondrial poly(A) polymerase |
| chr9_-_91100482 | 0.07 |
ENSRNOT00000064642
|
LOC100911860
|
ubiquitin carboxyl-terminal hydrolase 40-like |
| chr2_+_248398917 | 0.07 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr8_+_19888667 | 0.07 |
ENSRNOT00000078593
|
Zfp317
|
zinc finger protein 317 |
| chr5_-_16799776 | 0.07 |
ENSRNOT00000011721
|
Plag1
|
PLAG1 zinc finger |
| chr10_-_52290657 | 0.07 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr17_-_87826421 | 0.07 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr2_-_157035483 | 0.07 |
ENSRNOT00000076491
|
LOC304239
|
similar to RalA binding protein 1 |
| chr9_+_64434904 | 0.07 |
ENSRNOT00000075508
|
LOC100911305
|
methionine--tRNA ligase, mitochondrial-like |
| chr16_-_25192675 | 0.07 |
ENSRNOT00000032289
|
March1
|
membrane associated ring-CH-type finger 1 |
| chr17_+_78764506 | 0.07 |
ENSRNOT00000077953
|
Suv39h2
|
suppressor of variegation 3-9 homolog 2 |
| chr1_-_87825333 | 0.07 |
ENSRNOT00000072689
|
Zfp74
|
zinc finger protein 74 |
| chr8_-_102149912 | 0.07 |
ENSRNOT00000011263
|
RGD1309079
|
similar to Ab2-095 |
| chr8_-_21481735 | 0.07 |
ENSRNOT00000038069
|
Zfp426
|
zinc finger protein 426 |
| chr3_+_8430829 | 0.07 |
ENSRNOT00000090440
ENSRNOT00000060969 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr14_+_79205466 | 0.07 |
ENSRNOT00000085534
|
Tbc1d14
|
TBC1 domain family, member 14 |
| chr5_+_163867961 | 0.07 |
ENSRNOT00000081495
|
AABR07050399.1
|
|
| chr9_-_14550625 | 0.07 |
ENSRNOT00000078227
|
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr9_+_66888393 | 0.07 |
ENSRNOT00000023536
|
Carf
|
calcium responsive transcription factor |
| chr11_+_67101714 | 0.07 |
ENSRNOT00000078936
|
Cd86
|
CD86 molecule |
| chr13_-_82295123 | 0.07 |
ENSRNOT00000090120
|
LOC498265
|
similar to hypothetical protein FLJ10706 |
| chr10_-_34242985 | 0.07 |
ENSRNOT00000046438
|
RGD1559575
|
similar to novel protein |
| chr7_-_120770435 | 0.06 |
ENSRNOT00000077000
|
Ddx17
|
DEAD-box helicase 17 |
| chr18_-_43945273 | 0.06 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr11_-_14160697 | 0.06 |
ENSRNOT00000081096
|
Hspa13
|
heat shock protein 70 family, member 13 |
| chr18_-_26656879 | 0.06 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr15_-_29548400 | 0.06 |
ENSRNOT00000078176
|
AABR07017639.2
|
|
| chr2_+_220432037 | 0.06 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr4_-_176606382 | 0.06 |
ENSRNOT00000065576
|
Recql
|
RecQ like helicase |
| chr15_+_56666012 | 0.06 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr2_+_30664639 | 0.06 |
ENSRNOT00000076372
ENSRNOT00000076294 ENSRNOT00000076434 ENSRNOT00000076484 |
Taf9
|
TATA-box binding protein associated factor 9 |
| chr15_-_55424024 | 0.06 |
ENSRNOT00000077733
|
Nudt15
|
nudix hydrolase 15 |
| chr1_-_53802658 | 0.06 |
ENSRNOT00000032667
|
Afdn
|
afadin, adherens junction formation factor |
| chr6_+_8284878 | 0.06 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr7_-_107768072 | 0.06 |
ENSRNOT00000093189
|
Ndrg1
|
N-myc downstream regulated 1 |
| chr17_+_84881414 | 0.06 |
ENSRNOT00000034157
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr12_-_38916237 | 0.06 |
ENSRNOT00000074517
|
Tmem120b
|
transmembrane protein 120B |
| chr19_-_22194740 | 0.06 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr1_+_154446156 | 0.06 |
ENSRNOT00000093052
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chrX_+_33884499 | 0.06 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr4_-_51943519 | 0.05 |
ENSRNOT00000060459
|
Pot1
|
protection of telomeres 1 |
| chr18_+_30487264 | 0.05 |
ENSRNOT00000040125
|
Pcdhb10
|
protocadherin beta 10 |
| chr6_-_108076186 | 0.05 |
ENSRNOT00000014814
|
Fam161b
|
family with sequence similarity 161, member B |
| chrX_-_128268285 | 0.05 |
ENSRNOT00000009755
ENSRNOT00000081880 |
Thoc2
|
THO complex 2 |
| chr1_+_164706276 | 0.05 |
ENSRNOT00000024311
|
Olr36
|
olfactory receptor 36 |
| chrM_+_3904 | 0.05 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr5_+_36555109 | 0.05 |
ENSRNOT00000061128
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr6_-_3355339 | 0.05 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr7_-_124929025 | 0.05 |
ENSRNOT00000015447
|
Efcab6
|
EF-hand calcium binding domain 6 |
| chr2_-_30664163 | 0.05 |
ENSRNOT00000024801
|
Rad17
|
RAD17 checkpoint clamp loader component |
| chr1_+_61382118 | 0.05 |
ENSRNOT00000050119
|
Zfp52
|
zinc finger protein 52 |
| chr3_-_45169118 | 0.05 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr14_-_45859908 | 0.05 |
ENSRNOT00000086994
|
Pgm2
|
phosphoglucomutase 2 |
| chr1_-_64147251 | 0.05 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr19_-_49510901 | 0.05 |
ENSRNOT00000082929
ENSRNOT00000079969 |
LOC687399
|
hypothetical protein LOC687399 |
| chr14_+_39663421 | 0.05 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr19_+_46455761 | 0.05 |
ENSRNOT00000034012
ENSRNOT00000093392 |
Nudt7
|
nudix hydrolase 7 |
| chr4_+_176606649 | 0.05 |
ENSRNOT00000084700
|
Golt1b
|
golgi transport 1B |
| chr14_-_34561696 | 0.05 |
ENSRNOT00000059763
|
Srd5a3
|
steroid 5 alpha-reductase 3 |
| chr5_+_58995249 | 0.05 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chrX_+_77143892 | 0.05 |
ENSRNOT00000085227
|
Atp7a
|
ATPase copper transporting alpha |
| chr17_-_53713408 | 0.04 |
ENSRNOT00000022445
|
LOC100912163
|
AT-rich interactive domain-containing protein 4B-like |
| chr7_+_125189827 | 0.04 |
ENSRNOT00000080152
ENSRNOT00000082805 |
Parvb
|
parvin, beta |
| chr3_+_110442637 | 0.04 |
ENSRNOT00000010471
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr15_+_39779648 | 0.04 |
ENSRNOT00000084505
|
Cab39l
|
calcium binding protein 39-like |
| chr10_+_59000397 | 0.04 |
ENSRNOT00000021134
|
Mybbp1a
|
MYB binding protein 1a |
| chr18_-_4294136 | 0.04 |
ENSRNOT00000091909
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr8_+_64610952 | 0.04 |
ENSRNOT00000015963
|
Myo9a
|
myosin IXA |
| chr5_+_142986526 | 0.04 |
ENSRNOT00000012811
|
Rspo1
|
R-spondin 1 |
| chr19_-_37796089 | 0.04 |
ENSRNOT00000024891
|
Ranbp10
|
RAN binding protein 10 |
| chrX_+_73390903 | 0.04 |
ENSRNOT00000077002
|
Zfp449
|
zinc finger protein 449 |
| chrX_+_84064427 | 0.04 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr3_-_104018861 | 0.04 |
ENSRNOT00000008387
|
Chrm5
|
cholinergic receptor, muscarinic 5 |
| chr3_+_8547349 | 0.04 |
ENSRNOT00000079108
ENSRNOT00000091697 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr5_+_120568242 | 0.04 |
ENSRNOT00000050597
|
Lepr
|
leptin receptor |
| chr5_-_12172009 | 0.04 |
ENSRNOT00000061903
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr8_-_130338325 | 0.04 |
ENSRNOT00000071697
|
Sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr3_+_151688454 | 0.04 |
ENSRNOT00000026856
|
Romo1
|
reactive oxygen species modulator 1 |
| chr3_+_107142762 | 0.04 |
ENSRNOT00000006177
|
RGD1563680
|
similar to CDNA sequence BC052040 |
| chr13_-_76049363 | 0.04 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr5_-_93244202 | 0.04 |
ENSRNOT00000075474
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr14_+_2892753 | 0.04 |
ENSRNOT00000061630
|
Evi5
|
ecotropic viral integration site 5 |
| chr1_+_30863217 | 0.04 |
ENSRNOT00000015421
|
Rnf146
|
ring finger protein 146 |
| chr5_-_113532878 | 0.04 |
ENSRNOT00000010173
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr17_-_76281660 | 0.04 |
ENSRNOT00000058070
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr4_-_148437961 | 0.04 |
ENSRNOT00000082907
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr3_+_93909156 | 0.04 |
ENSRNOT00000090365
|
Lmo2
|
LIM domain only 2 |
| chr6_-_103935122 | 0.03 |
ENSRNOT00000058354
|
LOC500684
|
hypothetical protein LOC500684 |
| chr5_+_144106802 | 0.03 |
ENSRNOT00000035637
ENSRNOT00000079779 |
Lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr7_+_15247681 | 0.03 |
ENSRNOT00000006044
|
Zfp472
|
zinc finger protein 472 |
| chr13_+_43818010 | 0.03 |
ENSRNOT00000005532
|
LOC100909664
|
centrosomal protein of 170 kDa-like |
| chr4_-_66955732 | 0.03 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr4_-_150829913 | 0.03 |
ENSRNOT00000041571
|
Cacna1c
|
calcium voltage-gated channel subunit alpha1 C |
| chr12_-_5685448 | 0.03 |
ENSRNOT00000077167
|
Fry
|
FRY microtubule binding protein |
| chr4_-_150829741 | 0.03 |
ENSRNOT00000051846
ENSRNOT00000052017 |
Cacna1c
|
calcium voltage-gated channel subunit alpha1 C |
| chr9_+_11064605 | 0.03 |
ENSRNOT00000075121
|
Stap2
|
signal transducing adaptor family member 2 |
| chr12_+_41316764 | 0.03 |
ENSRNOT00000090867
|
Oas3
|
2'-5'-oligoadenylate synthetase 3 |
| chr1_-_67094567 | 0.03 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr1_+_201337416 | 0.03 |
ENSRNOT00000067742
|
Btbd16
|
BTB domain containing 16 |
| chr10_-_86645529 | 0.03 |
ENSRNOT00000011947
|
Med24
|
mediator complex subunit 24 |
| chr1_-_62114672 | 0.03 |
ENSRNOT00000072607
|
AABR07001923.1
|
|
| chr2_-_60461458 | 0.03 |
ENSRNOT00000024285
|
Brix1
|
BRX1, biogenesis of ribosomes |
| chr13_-_94859390 | 0.03 |
ENSRNOT00000005467
ENSRNOT00000079763 |
AABR07021840.1
|
|
| chr20_+_8312792 | 0.03 |
ENSRNOT00000083108
|
Cmtr1
|
cap methyltransferase 1 |
| chr2_-_35870578 | 0.03 |
ENSRNOT00000066069
|
Rnf180
|
ring finger protein 180 |
| chr1_-_167698263 | 0.03 |
ENSRNOT00000093046
|
Trim21
|
tripartite motif-containing 21 |
| chr1_-_126211439 | 0.03 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr18_-_6782757 | 0.03 |
ENSRNOT00000068150
|
Aqp4
|
aquaporin 4 |
| chr7_+_117456039 | 0.03 |
ENSRNOT00000051754
|
Mroh1
|
maestro heat-like repeat family member 1 |
| chr13_+_78979321 | 0.03 |
ENSRNOT00000003857
|
Ankrd45
|
ankyrin repeat domain 45 |
| chrX_+_123751089 | 0.03 |
ENSRNOT00000092384
|
Nkap
|
NFKB activating protein |
| chr8_-_40883880 | 0.03 |
ENSRNOT00000075593
|
LOC102550797
|
disks large homolog 5-like |
| chr2_-_187786700 | 0.03 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr6_+_76079880 | 0.03 |
ENSRNOT00000009304
|
RGD1305089
|
similar to 1110008L16Rik protein |
| chr8_+_127702534 | 0.03 |
ENSRNOT00000075793
|
Ctdspl
|
CTD small phosphatase like |
| chr5_-_113880911 | 0.03 |
ENSRNOT00000029441
|
Eqtn
|
equatorin |
| chr14_+_10786399 | 0.03 |
ENSRNOT00000003006
|
Lin54
|
lin-54 DREAM MuvB core complex component |
| chr9_-_11027506 | 0.03 |
ENSRNOT00000071107
|
Chaf1a
|
chromatin assembly factor 1 subunit A |
| chr20_+_49319307 | 0.03 |
ENSRNOT00000057078
|
Atg5
|
autophagy related 5 |
| chr20_+_20259254 | 0.03 |
ENSRNOT00000085985
ENSRNOT00000077620 ENSRNOT00000090273 |
Ank3
|
ankyrin 3 |
| chr18_-_44141865 | 0.03 |
ENSRNOT00000083407
|
LOC103694210
|
NACHT and WD repeat domain-containing protein 2-like |
| chr14_+_115396017 | 0.03 |
ENSRNOT00000067393
|
Asb3
|
ankyrin repeat and SOCS box-containing 3 |
| chr16_+_23447366 | 0.03 |
ENSRNOT00000068629
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_-_227441442 | 0.03 |
ENSRNOT00000028433
|
Ms4a1
|
membrane spanning 4-domains A1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Isl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006431 | methionyl-tRNA aminoacylation(GO:0006431) |
| 0.1 | 0.2 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 0.0 | 0.1 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:2000458 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0070662 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:1902962 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0060383 | positive regulation of DNA strand elongation(GO:0060383) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.0 | GO:0046356 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.0 | 0.2 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:0042262 | dGTP catabolic process(GO:0006203) DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.0 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.1 | GO:0032429 | negative regulation of phospholipase activity(GO:0010519) regulation of phospholipase A2 activity(GO:0032429) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.0 | GO:1904959 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.0 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004825 | methionine-tRNA ligase activity(GO:0004825) |
| 0.1 | 0.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.2 | GO:0070401 | NADP-retinol dehydrogenase activity(GO:0052650) NADP+ binding(GO:0070401) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0032551 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.0 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |