Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
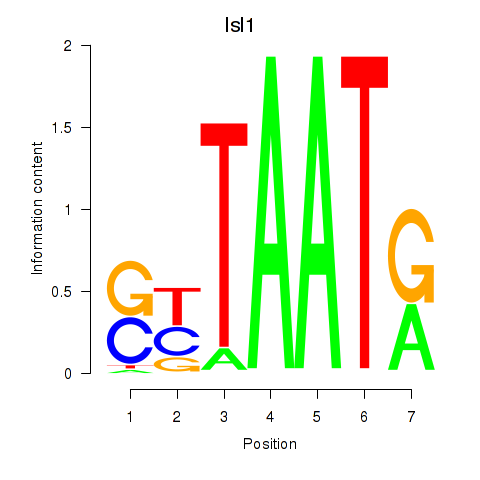
Results for Isl1
Z-value: 0.24
Transcription factors associated with Isl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Isl1
|
ENSRNOG00000012556 | ISL LIM homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isl1 | rn6_v1_chr2_-_48501436_48501436 | -0.37 | 5.4e-01 | Click! |
Activity profile of Isl1 motif
Sorted Z-values of Isl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_43638161 | 0.11 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr10_-_64657089 | 0.09 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chrX_+_153677811 | 0.08 |
ENSRNOT00000072808
|
AABR07042361.1
|
|
| chr19_-_58735173 | 0.07 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chr3_+_54253949 | 0.07 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr3_+_8192575 | 0.06 |
ENSRNOT00000060977
|
Rapgef1
|
Rap guanine nucleotide exchange factor 1 |
| chr20_-_12820466 | 0.06 |
ENSRNOT00000001699
|
Ftcd
|
formimidoyltransferase cyclodeaminase |
| chr2_+_66940057 | 0.06 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr15_+_62406873 | 0.05 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr9_-_52238564 | 0.05 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr15_-_45927804 | 0.05 |
ENSRNOT00000086271
|
Ints6
|
integrator complex subunit 6 |
| chr1_-_19376301 | 0.05 |
ENSRNOT00000015547
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr15_+_56666012 | 0.05 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr5_+_172364421 | 0.04 |
ENSRNOT00000018769
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr3_-_44177689 | 0.04 |
ENSRNOT00000006387
|
Cytip
|
cytohesin 1 interacting protein |
| chr15_+_4209703 | 0.04 |
ENSRNOT00000082236
|
Ppp3cb
|
protein phosphatase 3 catalytic subunit beta |
| chr3_-_25212049 | 0.04 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr14_+_69800156 | 0.04 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr3_+_33440191 | 0.04 |
ENSRNOT00000034632
ENSRNOT00000092907 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr8_-_45137893 | 0.04 |
ENSRNOT00000010743
|
RGD1309108
|
similar to hypothetical protein FLJ23554 |
| chr18_-_28017925 | 0.04 |
ENSRNOT00000075420
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr10_-_21265026 | 0.03 |
ENSRNOT00000011922
|
Tenm2
|
teneurin transmembrane protein 2 |
| chr10_-_31359699 | 0.03 |
ENSRNOT00000081280
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr1_-_88240057 | 0.03 |
ENSRNOT00000073092
|
LOC103690038
|
zinc finger protein 569 |
| chr18_-_4294136 | 0.03 |
ENSRNOT00000091909
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr20_+_48504264 | 0.03 |
ENSRNOT00000087740
|
Cdc40
|
cell division cycle 40 |
| chr2_-_53300404 | 0.03 |
ENSRNOT00000088876
|
Ghr
|
growth hormone receptor |
| chr17_-_84247038 | 0.03 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr6_-_80107236 | 0.03 |
ENSRNOT00000006369
|
Sec23a
|
Sec23 homolog A, coat complex II component |
| chr1_-_126211439 | 0.03 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr13_-_89242443 | 0.02 |
ENSRNOT00000029202
|
Atf6
|
activating transcription factor 6 |
| chrX_-_128268285 | 0.02 |
ENSRNOT00000009755
ENSRNOT00000081880 |
Thoc2
|
THO complex 2 |
| chr2_+_4989295 | 0.02 |
ENSRNOT00000041541
|
Fam172a
|
family with sequence similarity 172, member A |
| chr3_-_71798531 | 0.02 |
ENSRNOT00000088170
|
Calcrl
|
calcitonin receptor like receptor |
| chr17_+_5513930 | 0.02 |
ENSRNOT00000091370
|
Agtpbp1
|
ATP/GTP binding protein 1 |
| chr14_-_3288017 | 0.02 |
ENSRNOT00000080452
|
LOC689986
|
hypothetical protein LOC689986 |
| chr5_+_113725717 | 0.02 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr2_+_34312766 | 0.02 |
ENSRNOT00000060962
|
Cenpk
|
centromere protein K |
| chr12_+_37444072 | 0.02 |
ENSRNOT00000077380
ENSRNOT00000092699 ENSRNOT00000001373 ENSRNOT00000092416 |
Eif2b1
|
eukaryotic translation initiation factor 2B subunit 1 alpha |
| chr2_+_157453428 | 0.02 |
ENSRNOT00000051494
|
Lekr1
|
leucine, glutamate and lysine rich 1 |
| chr8_+_99894762 | 0.02 |
ENSRNOT00000081768
|
Plscr4
|
phospholipid scramblase 4 |
| chr15_-_52320385 | 0.02 |
ENSRNOT00000067776
|
Dmtn
|
dematin actin binding protein |
| chr20_+_9948908 | 0.02 |
ENSRNOT00000001541
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr13_-_102942863 | 0.02 |
ENSRNOT00000003198
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr1_-_255815733 | 0.02 |
ENSRNOT00000047387
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr1_+_239412884 | 0.01 |
ENSRNOT00000067384
|
Tmem2
|
transmembrane protein 2 |
| chr14_-_5033941 | 0.01 |
ENSRNOT00000072818
ENSRNOT00000082331 |
Zfp326
|
zinc finger protein 326 |
| chr14_-_19191863 | 0.01 |
ENSRNOT00000003921
|
Alb
|
albumin |
| chr11_-_17119582 | 0.01 |
ENSRNOT00000087665
|
RGD1563888
|
similar to DNA segment, Chr 16, ERATO Doi 472, expressed |
| chr2_+_83393282 | 0.01 |
ENSRNOT00000044871
|
Ctnnd2
|
catenin delta 2 |
| chr13_+_56513286 | 0.01 |
ENSRNOT00000015596
|
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr6_-_117972898 | 0.01 |
ENSRNOT00000032968
|
AABR07065265.1
|
|
| chrX_-_63182385 | 0.01 |
ENSRNOT00000076613
|
Zfx
|
zinc finger protein X-linked |
| chr4_+_157836912 | 0.01 |
ENSRNOT00000067271
|
Scnn1a
|
sodium channel epithelial 1 alpha subunit |
| chr7_-_134722215 | 0.01 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr7_+_138707426 | 0.01 |
ENSRNOT00000037874
|
Pced1b
|
PC-esterase domain containing 1B |
| chr2_+_188844073 | 0.01 |
ENSRNOT00000028117
|
Kcnn3
|
potassium calcium-activated channel subfamily N member 3 |
| chr4_+_57883849 | 0.01 |
ENSRNOT00000013880
|
Cpa4
|
carboxypeptidase A4 |
| chr8_+_57936650 | 0.01 |
ENSRNOT00000089686
|
Exph5
|
exophilin 5 |
| chr18_-_26656879 | 0.00 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr2_-_157759819 | 0.00 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr15_+_12827707 | 0.00 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr15_+_28023018 | 0.00 |
ENSRNOT00000090272
|
Rnase4
|
ribonuclease A family member 4 |
| chrX_+_23081125 | 0.00 |
ENSRNOT00000071639
|
AABR07037520.1
|
|
| chr8_+_64489942 | 0.00 |
ENSRNOT00000083666
|
Pkm
|
pyruvate kinase, muscle |
| chr6_-_107678156 | 0.00 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr20_+_48503973 | 0.00 |
ENSRNOT00000064081
|
Cdc40
|
cell division cycle 40 |
| chr5_+_18901039 | 0.00 |
ENSRNOT00000012066
|
Fam110b
|
family with sequence similarity 110, member B |
| chr14_-_86560694 | 0.00 |
ENSRNOT00000075827
|
Npc1l1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr3_-_161819029 | 0.00 |
ENSRNOT00000091834
|
LOC102555457
|
engulfment and cell motility protein 2-like |
| chr4_+_158088505 | 0.00 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr1_-_102826965 | 0.00 |
ENSRNOT00000078692
|
Saa4
|
serum amyloid A4 |
| chr2_-_100372252 | 0.00 |
ENSRNOT00000011890
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chrX_+_78769419 | 0.00 |
ENSRNOT00000003190
|
Tbx22
|
T-box 22 |
| chr4_-_7885301 | 0.00 |
ENSRNOT00000035034
|
Rint1
|
RAD50 interactor 1 |
| chr17_-_77527894 | 0.00 |
ENSRNOT00000032173
|
Bend7
|
BEN domain containing 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Isl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 0.0 | 0.0 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.0 | 0.0 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |