Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Irx4
Z-value: 0.19
Transcription factors associated with Irx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irx4
|
ENSRNOG00000012720 | iroquois homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irx4 | rn6_v1_chr1_-_32643771_32643771 | -0.81 | 9.7e-02 | Click! |
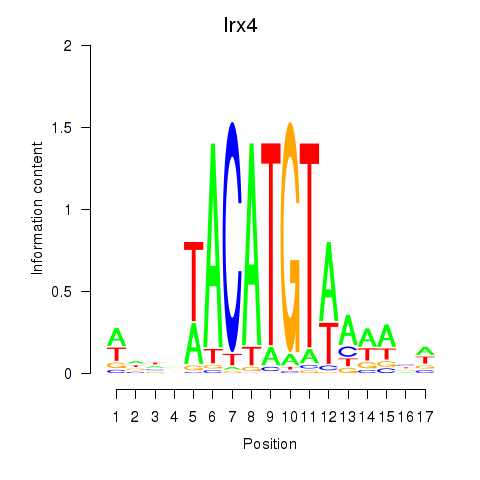
Activity profile of Irx4 motif
Sorted Z-values of Irx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_19854836 | 0.08 |
ENSRNOT00000075010
|
AABR07037395.2
|
|
| chr16_-_6404578 | 0.05 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr16_-_6405117 | 0.05 |
ENSRNOT00000047737
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chrX_-_158288109 | 0.04 |
ENSRNOT00000072527
|
LOC103690175
|
PHD finger protein 6 |
| chrX_+_18163358 | 0.04 |
ENSRNOT00000045517
|
LOC367746
|
similar to Spindlin-like protein 2 (SPIN-2) |
| chr17_-_51912496 | 0.03 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr16_-_6404957 | 0.02 |
ENSRNOT00000048459
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr7_-_24667301 | 0.02 |
ENSRNOT00000009154
|
Tmem263
|
transmembrane protein 263 |
| chr3_-_153246433 | 0.02 |
ENSRNOT00000067748
|
Samhd1
|
SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 |
| chr2_-_184289126 | 0.02 |
ENSRNOT00000081678
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr18_+_29490981 | 0.02 |
ENSRNOT00000032316
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chrX_-_45284341 | 0.01 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chr16_-_9658484 | 0.01 |
ENSRNOT00000065216
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr12_-_30491102 | 0.01 |
ENSRNOT00000001224
|
Sumf2
|
sulfatase modifying factor 2 |
| chr13_-_67819835 | 0.01 |
ENSRNOT00000093666
|
Hmcn1
|
hemicentin 1 |
| chr2_+_192538899 | 0.01 |
ENSRNOT00000045691
ENSRNOT00000085931 |
LOC102550416
|
small proline-rich protein 2I-like |
| chr4_-_28437676 | 0.01 |
ENSRNOT00000012995
|
Hepacam2
|
HEPACAM family member 2 |
| chrX_-_83864150 | 0.01 |
ENSRNOT00000073734
|
Obp1f
|
odorant binding protein I f |
| chr18_-_60835739 | 0.01 |
ENSRNOT00000039569
|
LOC291543
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr4_-_120559078 | 0.00 |
ENSRNOT00000085730
ENSRNOT00000079575 |
Kbtbd12
|
kelch repeat and BTB domain containing 12 |
| chr3_-_66885085 | 0.00 |
ENSRNOT00000084299
ENSRNOT00000090547 |
Pde1a
|
phosphodiesterase 1A |
| chr20_-_821332 | 0.00 |
ENSRNOT00000040957
|
Olr1692
|
olfactory receptor 1692 |
| chr1_+_82089922 | 0.00 |
ENSRNOT00000071251
|
Zfp526
|
zinc finger protein 526 |
| chr10_+_3179346 | 0.00 |
ENSRNOT00000082082
|
Rrn3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chr10_-_95431312 | 0.00 |
ENSRNOT00000085552
|
AABR07030603.2
|
|
| chr1_-_62681471 | 0.00 |
ENSRNOT00000042466
|
Vom2r15
|
vomeronasal 2 receptor, 15 |
| chr3_-_46601409 | 0.00 |
ENSRNOT00000079261
|
Pla2r1
|
phospholipase A2 receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |