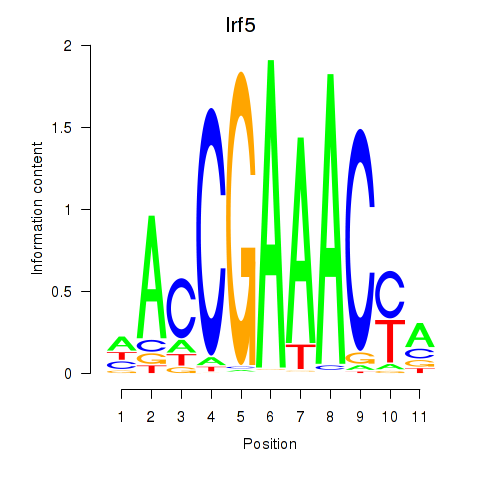
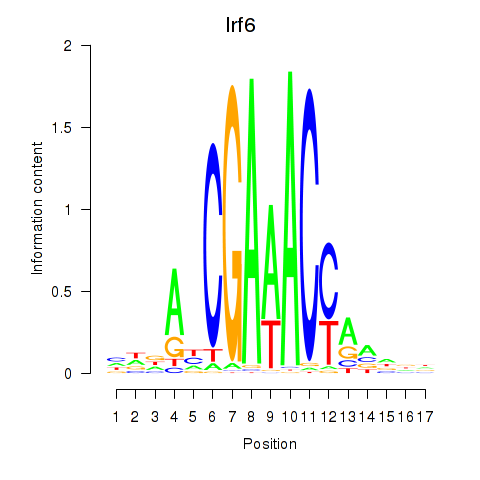
Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Irf5_Irf6
Z-value: 0.35
Transcription factors associated with Irf5_Irf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf5
|
ENSRNOG00000007437 | interferon regulatory factor 5 |
|
Irf6
|
ENSRNOG00000005082 | interferon regulatory factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf5 | rn6_v1_chr4_+_56805132_56805158 | 0.94 | 1.8e-02 | Click! |
| Irf6 | rn6_v1_chr13_+_111870121_111870121 | -0.68 | 2.0e-01 | Click! |
Activity profile of Irf5_Irf6 motif
Sorted Z-values of Irf5_Irf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_76343847 | 0.09 |
ENSRNOT00000055674
|
Trim25
|
tripartite motif-containing 25 |
| chr2_-_247988462 | 0.09 |
ENSRNOT00000022387
|
Pdlim5
|
PDZ and LIM domain 5 |
| chr5_-_56536772 | 0.08 |
ENSRNOT00000060765
|
Ddx58
|
DEXD/H-box helicase 58 |
| chr20_-_4542073 | 0.08 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr4_-_88649216 | 0.07 |
ENSRNOT00000058626
|
Herc6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr9_+_60039297 | 0.07 |
ENSRNOT00000016262
|
Slc39a10
|
solute carrier family 39 member 10 |
| chr6_+_43743702 | 0.06 |
ENSRNOT00000082570
|
Grhl1
|
grainyhead-like transcription factor 1 |
| chr2_-_30577218 | 0.06 |
ENSRNOT00000024674
|
Ocln
|
occludin |
| chr1_+_220948331 | 0.06 |
ENSRNOT00000036185
|
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr2_-_30576591 | 0.05 |
ENSRNOT00000084667
|
Ocln
|
occludin |
| chr14_+_113530470 | 0.05 |
ENSRNOT00000004919
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr1_+_247519939 | 0.05 |
ENSRNOT00000034421
|
Cd274
|
CD274 molecule |
| chr15_+_3996328 | 0.05 |
ENSRNOT00000088252
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr5_+_116421894 | 0.05 |
ENSRNOT00000080577
ENSRNOT00000086628 ENSRNOT00000004017 |
Nfia
|
nuclear factor I/A |
| chr10_+_59259955 | 0.05 |
ENSRNOT00000021816
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr1_-_193328371 | 0.04 |
ENSRNOT00000019304
ENSRNOT00000031770 ENSRNOT00000019309 |
Arhgap17
|
Rho GTPase activating protein 17 |
| chr8_+_103774358 | 0.04 |
ENSRNOT00000014481
|
Xrn1
|
5'-3' exoribonuclease 1 |
| chr15_+_3996048 | 0.04 |
ENSRNOT00000078653
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr10_-_82027119 | 0.04 |
ENSRNOT00000003914
|
Luc7l3
|
LUC7-like 3 pre-mRNA splicing factor |
| chr3_-_37445907 | 0.03 |
ENSRNOT00000006172
|
Rbm43
|
RNA binding motif protein 43 |
| chr9_-_44560837 | 0.03 |
ENSRNOT00000090870
|
Mitd1
|
microtubule interacting and trafficking domain containing 1 |
| chr11_+_68105369 | 0.03 |
ENSRNOT00000046888
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr9_-_54327958 | 0.03 |
ENSRNOT00000019465
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr11_+_67757928 | 0.03 |
ENSRNOT00000039215
|
Dtx3l
|
deltex E3 ubiquitin ligase 3L |
| chr15_+_60084918 | 0.03 |
ENSRNOT00000012632
|
Epsti1
|
epithelial stromal interaction 1 |
| chr17_+_36334147 | 0.03 |
ENSRNOT00000050261
|
E2f3
|
E2F transcription factor 3 |
| chr2_+_196120580 | 0.03 |
ENSRNOT00000068528
|
Rfx5
|
regulatory factor X5 |
| chr20_-_47910375 | 0.03 |
ENSRNOT00000000348
|
Sobp
|
sine oculis binding protein homolog |
| chr9_-_44483655 | 0.02 |
ENSRNOT00000024959
|
Mitd1
|
microtubule interacting and trafficking domain containing 1 |
| chr10_-_54467956 | 0.02 |
ENSRNOT00000065383
|
Usp43
|
ubiquitin specific peptidase 43 |
| chr6_-_51257625 | 0.02 |
ENSRNOT00000012004
|
Hbp1
|
HMG-box transcription factor 1 |
| chr3_+_8453609 | 0.02 |
ENSRNOT00000041921
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr1_-_220938814 | 0.02 |
ENSRNOT00000028081
|
Ovol1
|
ovo like transcriptional repressor 1 |
| chr6_+_125870372 | 0.02 |
ENSRNOT00000008612
|
Cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr6_+_134958854 | 0.02 |
ENSRNOT00000073950
ENSRNOT00000009377 |
Dync1h1
|
dynein cytoplasmic 1 heavy chain 1 |
| chrX_-_104493714 | 0.02 |
ENSRNOT00000064458
ENSRNOT00000080386 |
Pcdh19
|
protocadherin 19 |
| chr10_-_4257868 | 0.02 |
ENSRNOT00000035886
|
Tnfrsf17
|
TNF receptor superfamily member 17 |
| chr3_-_29984201 | 0.01 |
ENSRNOT00000006350
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr9_-_75528644 | 0.01 |
ENSRNOT00000019283
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chrX_+_123751089 | 0.01 |
ENSRNOT00000092384
|
Nkap
|
NFKB activating protein |
| chr4_-_129544429 | 0.01 |
ENSRNOT00000091507
ENSRNOT00000083461 ENSRNOT00000087215 |
Tmf1
|
TATA element modulatory factor 1 |
| chrX_+_123751293 | 0.01 |
ENSRNOT00000089883
|
Nkap
|
NFKB activating protein |
| chr8_-_132911193 | 0.01 |
ENSRNOT00000087799
|
Fyco1
|
FYVE and coiled-coil domain containing 1 |
| chrX_+_44830849 | 0.01 |
ENSRNOT00000079680
|
Tbl1x
|
transducin (beta)-like 1 X-linked |
| chr20_+_3979035 | 0.01 |
ENSRNOT00000000529
|
Tap1
|
transporter 1, ATP binding cassette subfamily B member |
| chr6_-_1466201 | 0.01 |
ENSRNOT00000089185
|
Eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr10_-_68926264 | 0.01 |
ENSRNOT00000008661
|
Phb-ps1
|
prohibitin, pseudogene 1 |
| chr12_-_46414434 | 0.01 |
ENSRNOT00000041281
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chrX_-_64702441 | 0.01 |
ENSRNOT00000051132
|
Amer1
|
APC membrane recruitment protein 1 |
| chr11_+_45888221 | 0.01 |
ENSRNOT00000071932
ENSRNOT00000066008 ENSRNOT00000060802 |
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr10_+_46940965 | 0.01 |
ENSRNOT00000005366
|
Llgl1
|
LLGL1, scribble cell polarity complex component |
| chr16_-_19349080 | 0.01 |
ENSRNOT00000038494
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr10_+_13000090 | 0.00 |
ENSRNOT00000004845
|
Cldn6
|
claudin 6 |
| chr6_-_10592454 | 0.00 |
ENSRNOT00000020600
|
Pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr15_+_34270648 | 0.00 |
ENSRNOT00000026333
|
Rnf31
|
ring finger protein 31 |
| chr10_-_82963919 | 0.00 |
ENSRNOT00000005807
|
Dlx4
|
distal-less homeobox 4 |
| chr2_+_185393441 | 0.00 |
ENSRNOT00000015802
|
Sh3d19
|
SH3 domain containing 19 |
| chr8_+_126411829 | 0.00 |
ENSRNOT00000013450
|
Azi2
|
5-azacytidine induced 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf5_Irf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0000962 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |