Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
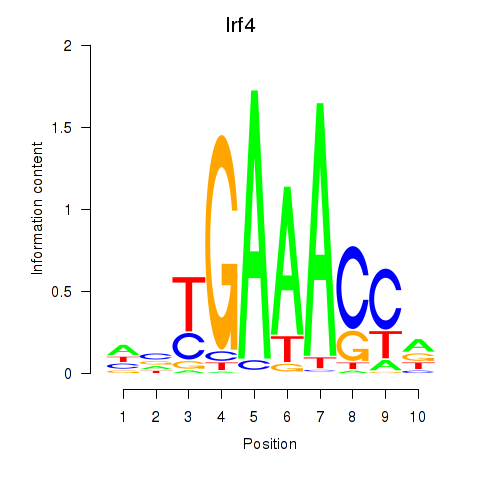
Results for Irf4
Z-value: 0.33
Transcription factors associated with Irf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf4
|
ENSRNOG00000061070 | interferon regulatory factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf4 | rn6_v1_chr17_-_34905117_34905117 | -0.55 | 3.3e-01 | Click! |
Activity profile of Irf4 motif
Sorted Z-values of Irf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_198008893 | 0.17 |
ENSRNOT00000025950
|
Il27
|
interleukin 27 |
| chr17_+_72429618 | 0.10 |
ENSRNOT00000026187
|
Gata3
|
GATA binding protein 3 |
| chr15_-_108898703 | 0.08 |
ENSRNOT00000067577
|
Zic5
|
Zic family member 5 |
| chr8_+_55050284 | 0.07 |
ENSRNOT00000013242
|
Pih1d2
|
PIH1 domain containing 2 |
| chr10_-_105412627 | 0.07 |
ENSRNOT00000034817
|
Qrich2
|
glutamine rich 2 |
| chr15_+_24141651 | 0.07 |
ENSRNOT00000082304
|
Lgals3
|
galectin 3 |
| chr1_+_220948331 | 0.07 |
ENSRNOT00000036185
|
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr17_+_47662532 | 0.05 |
ENSRNOT00000081314
|
Stard3nl
|
STARD3 N-terminal like |
| chr1_-_214252456 | 0.05 |
ENSRNOT00000023504
|
Irf7
|
interferon regulatory factor 7 |
| chr9_-_64096265 | 0.05 |
ENSRNOT00000013502
|
Tyw5
|
tRNA-yW synthesizing protein 5 |
| chr8_-_92942076 | 0.05 |
ENSRNOT00000056937
|
Fam46a
|
family with sequence similarity 46, member A |
| chr9_+_18001105 | 0.05 |
ENSRNOT00000071814
|
AABR07066826.2
|
|
| chr7_+_122700854 | 0.04 |
ENSRNOT00000086603
|
Rbx1
|
ring-box 1 |
| chr2_-_96032722 | 0.04 |
ENSRNOT00000015746
|
LOC100360846
|
proteasome subunit beta type 6-like |
| chr2_-_185852759 | 0.04 |
ENSRNOT00000049461
|
Mab21l2
|
mab-21 like 2 |
| chr9_+_84410972 | 0.04 |
ENSRNOT00000019724
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr20_-_50564987 | 0.04 |
ENSRNOT00000034485
ENSRNOT00000064466 |
Lin28b
|
lin-28 homolog B |
| chr6_-_135049728 | 0.04 |
ENSRNOT00000009556
|
Hsp90aa1
|
heat shock protein 90 alpha family class A member 1 |
| chr8_-_55050194 | 0.04 |
ENSRNOT00000013197
|
LOC100151767
|
hypothetical LOC100151767 |
| chr1_+_80056755 | 0.04 |
ENSRNOT00000021221
|
Snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr2_+_58724855 | 0.04 |
ENSRNOT00000089609
|
Capsl
|
calcyphosine-like |
| chr1_+_266333440 | 0.04 |
ENSRNOT00000071243
|
Sfxn2
|
sideroflexin 2 |
| chr1_+_196095214 | 0.03 |
ENSRNOT00000080741
|
LOC691716
|
similar to ribosomal protein S15a |
| chrX_+_31984612 | 0.03 |
ENSRNOT00000005181
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr2_+_95077577 | 0.03 |
ENSRNOT00000046368
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr13_-_102643223 | 0.03 |
ENSRNOT00000003155
|
Hlx
|
H2.0-like homeobox |
| chr8_-_53223804 | 0.03 |
ENSRNOT00000008965
|
Htr3a
|
5-hydroxytryptamine receptor 3A |
| chr15_+_105640097 | 0.03 |
ENSRNOT00000014300
|
Mbnl2
|
muscleblind-like splicing regulator 2 |
| chrX_-_105308554 | 0.03 |
ENSRNOT00000045556
|
Taf7l
|
TATA-box binding protein associated factor 7-like |
| chr15_+_34256071 | 0.03 |
ENSRNOT00000025887
|
Psme1
|
proteasome activator subunit 1 |
| chrM_+_8599 | 0.03 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr15_+_43905099 | 0.03 |
ENSRNOT00000016568
|
Ebf2
|
early B-cell factor 2 |
| chr8_+_117307339 | 0.03 |
ENSRNOT00000064571
|
Qrich1
|
glutamine-rich 1 |
| chr7_-_143353925 | 0.03 |
ENSRNOT00000068533
|
Krt71
|
keratin 71 |
| chr2_-_196046311 | 0.03 |
ENSRNOT00000028484
|
Psmb4
|
proteasome subunit beta 4 |
| chr17_-_32015071 | 0.03 |
ENSRNOT00000070876
|
LOC100911107
|
leukocyte elastase inhibitor A-like |
| chr10_+_74724549 | 0.03 |
ENSRNOT00000009077
|
Tex14
|
testis expressed 14, intercellular bridge forming factor |
| chr5_-_151898022 | 0.03 |
ENSRNOT00000000133
|
Pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr1_-_16687817 | 0.02 |
ENSRNOT00000091376
ENSRNOT00000081620 |
Myb
|
MYB proto-oncogene, transcription factor |
| chr16_+_2706428 | 0.02 |
ENSRNOT00000077117
|
Il17rd
|
interleukin 17 receptor D |
| chr2_-_210991509 | 0.02 |
ENSRNOT00000087329
|
Cyb561d1
|
cytochrome b561 family, member D1 |
| chr20_+_25990656 | 0.02 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr2_-_30577218 | 0.02 |
ENSRNOT00000024674
|
Ocln
|
occludin |
| chr13_-_79899479 | 0.02 |
ENSRNOT00000035815
|
RGD1309106
|
similar to hypothetical protein |
| chr6_+_147315328 | 0.02 |
ENSRNOT00000056654
|
Macc1
|
MACC1, MET transcriptional regulator |
| chr14_+_113530470 | 0.02 |
ENSRNOT00000004919
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr9_+_73493027 | 0.02 |
ENSRNOT00000074427
ENSRNOT00000089478 |
Unc80
|
unc-80 homolog, NALCN activator |
| chr5_-_107857320 | 0.02 |
ENSRNOT00000008898
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B |
| chr19_-_49448072 | 0.02 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr1_-_142183884 | 0.02 |
ENSRNOT00000016032
|
Fes
|
FES proto-oncogene, tyrosine kinase |
| chr13_+_48790951 | 0.02 |
ENSRNOT00000090233
|
Elk4
|
ELK4, ETS transcription factor |
| chr20_-_4698718 | 0.02 |
ENSRNOT00000047527
|
RT1-CE7
|
RT1 class I, locus CE7 |
| chr20_-_27682861 | 0.02 |
ENSRNOT00000057317
|
Fam26f
|
family with sequence similarity 26, member F |
| chr15_+_18710492 | 0.02 |
ENSRNOT00000012532
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr10_+_34277993 | 0.02 |
ENSRNOT00000055872
ENSRNOT00000003343 |
Ifi47
|
interferon gamma inducible protein 47 |
| chr18_+_83471342 | 0.02 |
ENSRNOT00000019384
|
Neto1
|
neuropilin and tolloid like 1 |
| chr2_-_30576591 | 0.02 |
ENSRNOT00000084667
|
Ocln
|
occludin |
| chr18_-_56728185 | 0.02 |
ENSRNOT00000066048
|
Ppargc1b
|
PPARG coactivator 1 beta |
| chr9_+_64095978 | 0.02 |
ENSRNOT00000021533
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr5_-_126782726 | 0.02 |
ENSRNOT00000048784
|
Tceanc2
|
transcription elongation factor A N-terminal and central domain containing 2 |
| chr3_-_52510507 | 0.02 |
ENSRNOT00000091259
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr7_+_91832988 | 0.02 |
ENSRNOT00000006410
|
Slc30a8
|
solute carrier family 30 member 8 |
| chr20_-_35450513 | 0.02 |
ENSRNOT00000087342
|
Man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr6_+_123974798 | 0.01 |
ENSRNOT00000075794
|
Kcnk13
|
potassium two pore domain channel subfamily K member 13 |
| chr1_-_47213749 | 0.01 |
ENSRNOT00000024656
|
Dynlt1
|
dynein light chain Tctex-type 1 |
| chr1_-_40921508 | 0.01 |
ENSRNOT00000026433
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr5_-_33892462 | 0.01 |
ENSRNOT00000009334
|
Atp6v0d2
|
ATPase H+ transporting V0 subunit D2 |
| chr4_+_112773938 | 0.01 |
ENSRNOT00000009321
|
Eva1a
|
eva-1 homolog A, regulator of programmed cell death |
| chr2_+_165076607 | 0.01 |
ENSRNOT00000012831
|
Il12a
|
interleukin 12A |
| chr5_-_59913348 | 0.01 |
ENSRNOT00000018164
|
LOC100359916
|
heterogeneous nuclear ribonucleoprotein K-like |
| chr5_-_107858104 | 0.01 |
ENSRNOT00000092196
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B |
| chr5_+_154077944 | 0.01 |
ENSRNOT00000031512
|
Il22ra1
|
interleukin 22 receptor subunit alpha 1 |
| chr1_+_260289589 | 0.01 |
ENSRNOT00000051058
|
Dntt
|
DNA nucleotidylexotransferase |
| chr11_+_67757928 | 0.01 |
ENSRNOT00000039215
|
Dtx3l
|
deltex E3 ubiquitin ligase 3L |
| chr15_-_9086282 | 0.01 |
ENSRNOT00000008989
|
Thrb
|
thyroid hormone receptor beta |
| chr3_+_120726906 | 0.01 |
ENSRNOT00000051069
|
Bcl2l11
|
BCL2 like 11 |
| chr1_-_201110928 | 0.01 |
ENSRNOT00000027721
|
Nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr20_-_4542073 | 0.01 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr17_+_36334147 | 0.01 |
ENSRNOT00000050261
|
E2f3
|
E2F transcription factor 3 |
| chr4_+_161685258 | 0.01 |
ENSRNOT00000008012
ENSRNOT00000008003 |
Foxm1
|
forkhead box M1 |
| chr7_-_15364203 | 0.01 |
ENSRNOT00000036473
|
RGD1310212
|
similar to KIAA1111-like protein |
| chr3_-_37445907 | 0.01 |
ENSRNOT00000006172
|
Rbm43
|
RNA binding motif protein 43 |
| chr8_-_40122450 | 0.01 |
ENSRNOT00000039729
|
Tbrg1
|
transforming growth factor beta regulator 1 |
| chr9_-_82514399 | 0.01 |
ENSRNOT00000026799
|
Dnpep
|
aspartyl aminopeptidase |
| chr12_-_47482961 | 0.01 |
ENSRNOT00000001600
|
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr1_-_266333105 | 0.01 |
ENSRNOT00000027093
|
Arl3
|
ADP ribosylation factor like GTPase 3 |
| chr1_+_13261876 | 0.00 |
ENSRNOT00000090703
|
Reps1
|
RALBP1 associated Eps domain containing 1 |
| chr7_-_24885775 | 0.00 |
ENSRNOT00000010024
|
Tcp11l2
|
t-complex 11 like 2 |
| chr15_+_3996328 | 0.00 |
ENSRNOT00000088252
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr2_-_57196568 | 0.00 |
ENSRNOT00000060146
|
Wdr70
|
WD repeat domain 70 |
| chr2_-_30633857 | 0.00 |
ENSRNOT00000085488
|
Marveld2
|
MARVEL domain containing 2 |
| chr5_-_62153762 | 0.00 |
ENSRNOT00000066001
ENSRNOT00000086962 |
Trim14
|
tripartite motif-containing 14 |
| chrX_+_85338928 | 0.00 |
ENSRNOT00000074811
ENSRNOT00000006907 |
Dach2
|
dachshund family transcription factor 2 |
| chr12_-_9998779 | 0.00 |
ENSRNOT00000001265
|
Rpl21
|
ribosomal protein L21 |
| chr15_+_3996048 | 0.00 |
ENSRNOT00000078653
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr17_-_69827112 | 0.00 |
ENSRNOT00000023835
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr10_-_89454681 | 0.00 |
ENSRNOT00000028109
|
Brca1
|
BRCA1, DNA repair associated |
| chr5_-_153924896 | 0.00 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr1_-_79930263 | 0.00 |
ENSRNOT00000019219
|
Foxa3
|
forkhead box A3 |
| chr20_+_25990304 | 0.00 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr10_+_76343847 | 0.00 |
ENSRNOT00000055674
|
Trim25
|
tripartite motif-containing 25 |
| chr13_+_82438697 | 0.00 |
ENSRNOT00000003759
|
Selp
|
selectin P |
| chr18_-_53181503 | 0.00 |
ENSRNOT00000066548
|
Fbn2
|
fibrillin 2 |
| chr6_+_135866739 | 0.00 |
ENSRNOT00000013460
|
Exoc3l4
|
exocyst complex component 3-like 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.1 | GO:1903769 | negative regulation of cell proliferation in bone marrow(GO:1903769) |
| 0.0 | 0.2 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) advanced glycation end-product receptor activity(GO:0050785) |
| 0.0 | 0.0 | GO:0002134 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |