Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
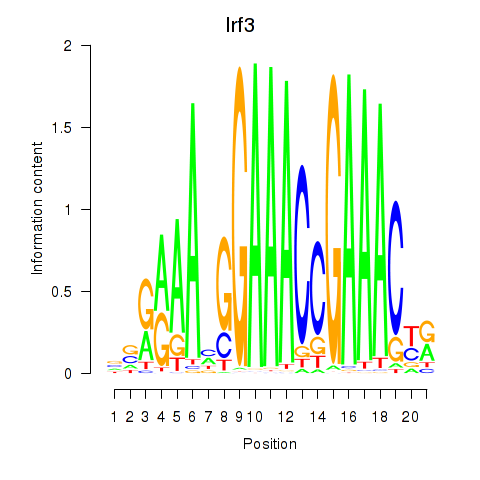
Results for Irf3
Z-value: 0.53
Transcription factors associated with Irf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf3
|
ENSRNOG00000043388 | interferon regulatory factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf3 | rn6_v1_chr1_+_100992405_100992405 | -0.02 | 9.7e-01 | Click! |
Activity profile of Irf3 motif
Sorted Z-values of Irf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_70744315 | 0.24 |
ENSRNOT00000014865
|
Ccl5
|
C-C motif chemokine ligand 5 |
| chr1_-_214252456 | 0.17 |
ENSRNOT00000023504
|
Irf7
|
interferon regulatory factor 7 |
| chr1_-_198008893 | 0.16 |
ENSRNOT00000025950
|
Il27
|
interleukin 27 |
| chr2_-_256915563 | 0.14 |
ENSRNOT00000084873
ENSRNOT00000029990 |
Ifi44
|
interferon-induced protein 44 |
| chr20_+_3979035 | 0.13 |
ENSRNOT00000000529
|
Tap1
|
transporter 1, ATP binding cassette subfamily B member |
| chr6_-_127337791 | 0.13 |
ENSRNOT00000032015
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr12_+_41200718 | 0.13 |
ENSRNOT00000038426
ENSRNOT00000048450 ENSRNOT00000067176 |
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr5_-_173626248 | 0.11 |
ENSRNOT00000039263
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr7_-_28040510 | 0.11 |
ENSRNOT00000005674
|
Ascl1
|
achaete-scute family bHLH transcription factor 1 |
| chr16_-_19942343 | 0.09 |
ENSRNOT00000087162
ENSRNOT00000091906 |
Bst2
|
bone marrow stromal cell antigen 2 |
| chr10_-_15459897 | 0.09 |
ENSRNOT00000027518
|
Decr2
|
2,4-dienoyl-CoA reductase 2 |
| chr9_-_81566642 | 0.09 |
ENSRNOT00000080345
|
Aamp
|
angio-associated, migratory cell protein |
| chr2_+_248249468 | 0.08 |
ENSRNOT00000022648
|
Gbp4
|
guanylate binding protein 4 |
| chr20_-_3978845 | 0.08 |
ENSRNOT00000000532
|
Psmb9
|
proteasome subunit beta 9 |
| chr15_+_34256071 | 0.08 |
ENSRNOT00000025887
|
Psme1
|
proteasome activator subunit 1 |
| chr19_-_37912027 | 0.07 |
ENSRNOT00000026462
|
Psmb10
|
proteasome subunit beta 10 |
| chr9_-_44483655 | 0.07 |
ENSRNOT00000024959
|
Mitd1
|
microtubule interacting and trafficking domain containing 1 |
| chr15_+_31948035 | 0.07 |
ENSRNOT00000071627
|
AABR07017868.1
|
|
| chr10_+_89358376 | 0.07 |
ENSRNOT00000028067
|
Ifi35
|
interferon-induced protein 35 |
| chr10_-_88611105 | 0.06 |
ENSRNOT00000024718
|
Dhx58
|
DEXH-box helicase 58 |
| chr4_-_66899914 | 0.06 |
ENSRNOT00000011481
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr1_+_169433539 | 0.06 |
ENSRNOT00000055210
|
Trim34
|
tripartite motif-containing 34 |
| chr11_+_68105369 | 0.06 |
ENSRNOT00000046888
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr15_-_34269851 | 0.05 |
ENSRNOT00000026279
|
Psme2
|
proteasome activator subunit 2 |
| chr3_+_114087287 | 0.05 |
ENSRNOT00000023017
|
B2m
|
beta-2 microglobulin |
| chr20_+_3269284 | 0.05 |
ENSRNOT00000089044
|
RT1-T24-1
|
RT1 class I, locus T24, gene 1 |
| chr10_-_34221928 | 0.05 |
ENSRNOT00000045545
|
Irgm
|
immunity-related GTPase M |
| chr1_-_227505267 | 0.05 |
ENSRNOT00000028443
|
Ms4a7
|
membrane spanning 4-domains A7 |
| chr4_-_61720956 | 0.04 |
ENSRNOT00000012879
|
Akr1b1
|
aldo-keto reductase family 1 member B |
| chr17_-_14637832 | 0.04 |
ENSRNOT00000074323
|
LOC103690116
|
osteomodulin |
| chr11_-_37914983 | 0.04 |
ENSRNOT00000039876
|
Mx1
|
myxovirus (influenza virus) resistance 1 |
| chr10_-_86144880 | 0.04 |
ENSRNOT00000067901
|
Med1
|
mediator complex subunit 1 |
| chr8_-_52828134 | 0.04 |
ENSRNOT00000007736
|
Rbm7
|
RNA binding motif protein 7 |
| chr11_+_67757928 | 0.04 |
ENSRNOT00000039215
|
Dtx3l
|
deltex E3 ubiquitin ligase 3L |
| chr3_-_171286413 | 0.04 |
ENSRNOT00000008365
ENSRNOT00000081036 |
Zbp1
|
Z-DNA binding protein 1 |
| chr9_+_81968332 | 0.04 |
ENSRNOT00000023152
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr17_-_6244612 | 0.03 |
ENSRNOT00000042145
ENSRNOT00000090914 ENSRNOT00000082611 |
Ntrk2
|
neurotrophic receptor tyrosine kinase 2 |
| chr3_-_151363115 | 0.03 |
ENSRNOT00000073492
|
Eif6
|
eukaryotic translation initiation factor 6 |
| chr3_-_37445907 | 0.03 |
ENSRNOT00000006172
|
Rbm43
|
RNA binding motif protein 43 |
| chr1_+_101517714 | 0.03 |
ENSRNOT00000028423
|
Plekha4
|
pleckstrin homology domain containing A4 |
| chr20_+_3242470 | 0.03 |
ENSRNOT00000078110
ENSRNOT00000041502 |
RT1-T24-2
RT1-T24-1
|
RT1 class I, locus T24, gene 2 RT1 class I, locus T24, gene 1 |
| chr17_-_15478343 | 0.03 |
ENSRNOT00000078029
|
Omd
|
osteomodulin |
| chr4_-_87111025 | 0.03 |
ENSRNOT00000018442
|
Kbtbd2
|
kelch repeat and BTB domain containing 2 |
| chr8_-_70215719 | 0.03 |
ENSRNOT00000015598
|
Rab11a
|
RAB11a, member RAS oncogene family |
| chr6_-_127653124 | 0.03 |
ENSRNOT00000047324
|
Serpina11
|
serpin family A member 11 |
| chr3_-_147809658 | 0.03 |
ENSRNOT00000085995
ENSRNOT00000045314 |
Rbck1
|
RANBP2-type and C3HC4-type zinc finger containing 1 |
| chr1_-_164101578 | 0.02 |
ENSRNOT00000022176
|
Uvrag
|
UV radiation resistance associated |
| chr13_+_29839867 | 0.02 |
ENSRNOT00000090623
|
AABR07020537.1
|
|
| chr1_-_197590277 | 0.02 |
ENSRNOT00000022502
ENSRNOT00000043433 |
Xpo6
|
exportin 6 |
| chr9_+_81566074 | 0.02 |
ENSRNOT00000074131
ENSRNOT00000046229 ENSRNOT00000090383 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr3_-_119405300 | 0.02 |
ENSRNOT00000015568
|
Sppl2a
|
signal peptide peptidase-like 2A |
| chrX_+_135187468 | 0.02 |
ENSRNOT00000006805
|
Bcorl1
|
BCL6 co-repressor-like 1 |
| chr2_+_1410934 | 0.02 |
ENSRNOT00000013625
ENSRNOT00000080222 |
Erap1
|
endoplasmic reticulum aminopeptidase 1 |
| chr9_-_44560837 | 0.02 |
ENSRNOT00000090870
|
Mitd1
|
microtubule interacting and trafficking domain containing 1 |
| chr4_+_67165883 | 0.02 |
ENSRNOT00000012280
|
Rab19
|
RAB19, member RAS oncogene family |
| chr3_+_47578384 | 0.02 |
ENSRNOT00000082769
|
Psmd14
|
proteasome 26S subunit, non-ATPase 14 |
| chr20_-_4677034 | 0.02 |
ENSRNOT00000075244
|
Rn60_20_0047.3
|
|
| chr20_+_3990820 | 0.02 |
ENSRNOT00000000528
|
Psmb8
|
proteasome subunit beta 8 |
| chr2_-_231877307 | 0.02 |
ENSRNOT00000073860
|
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr12_+_42343123 | 0.02 |
ENSRNOT00000043279
|
Oas1i
|
2 ' -5 ' oligoadenylate synthetase 1I |
| chr4_+_62380914 | 0.02 |
ENSRNOT00000029845
|
Tmem140
|
transmembrane protein 140 |
| chr13_+_71192142 | 0.01 |
ENSRNOT00000032157
|
Rnasel
|
ribonuclease L |
| chr11_-_70207361 | 0.01 |
ENSRNOT00000002444
|
Muc13
|
mucin 13, cell surface associated |
| chr3_-_163814799 | 0.01 |
ENSRNOT00000055127
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr16_+_75744786 | 0.01 |
ENSRNOT00000032368
|
Xkr5
|
XK related 5 |
| chr1_-_263511505 | 0.01 |
ENSRNOT00000023215
|
Cox15
|
COX15 cytochrome c oxidase assembly homolog |
| chr19_-_19377492 | 0.01 |
ENSRNOT00000080964
|
Nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr7_-_118933812 | 0.01 |
ENSRNOT00000031951
|
Apol9a
|
apolipoprotein L 9a |
| chr18_+_55576239 | 0.01 |
ENSRNOT00000050063
|
LOC100910979
|
interferon-inducible GTPase 1-like |
| chr3_-_48604097 | 0.01 |
ENSRNOT00000009620
|
Ifih1
|
interferon induced with helicase C domain 1 |
| chr17_+_52755621 | 0.01 |
ENSRNOT00000089943
|
Psme1-ps1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha), pseudogene 1 |
| chr8_-_23146689 | 0.01 |
ENSRNOT00000092200
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr4_-_150485781 | 0.01 |
ENSRNOT00000008763
|
Zfp248
|
zinc finger protein 248 |
| chr8_-_63034226 | 0.00 |
ENSRNOT00000043434
|
Pml
|
promyelocytic leukemia |
| chr16_+_6779281 | 0.00 |
ENSRNOT00000022865
|
Sfmbt1
|
Scm-like with four mbt domains 1 |
| chr12_+_41155497 | 0.00 |
ENSRNOT00000041741
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr4_-_163890801 | 0.00 |
ENSRNOT00000081946
|
Ly49si1
|
immunoreceptor Ly49si1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042509 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:0046968 | cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 0.0 | 0.1 | GO:0021550 | neuroblast differentiation(GO:0014016) spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) medulla oblongata development(GO:0021550) musculoskeletal movement, spinal reflex action(GO:0050883) olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) |
| 0.0 | 0.2 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.1 | GO:0002477 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:1901423 | response to benzene(GO:1901423) |
| 0.0 | 0.2 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.0 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.0 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |