Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
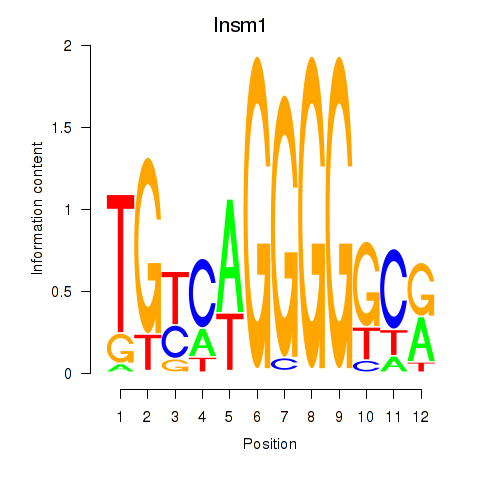
Results for Insm1
Z-value: 0.28
Transcription factors associated with Insm1
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Insm1 motif
Sorted Z-values of Insm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_127081978 | 0.14 |
ENSRNOT00000022945
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr18_-_55916220 | 0.12 |
ENSRNOT00000025934
|
Synpo
|
synaptopodin |
| chr4_+_113935492 | 0.12 |
ENSRNOT00000035329
|
LOC103692170
|
coiled-coil domain-containing protein 142 |
| chr5_+_142875773 | 0.11 |
ENSRNOT00000082120
ENSRNOT00000056496 |
Epha10
|
EPH receptor A10 |
| chr18_-_57114545 | 0.09 |
ENSRNOT00000026495
|
Afap1l1
|
actin filament associated protein 1-like 1 |
| chr6_+_119519714 | 0.08 |
ENSRNOT00000004955
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr13_-_48284408 | 0.07 |
ENSRNOT00000085967
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr1_+_165382690 | 0.07 |
ENSRNOT00000023802
|
C2cd3
|
C2 calcium-dependent domain containing 3 |
| chr6_+_26537707 | 0.07 |
ENSRNOT00000050102
|
Zfp513
|
zinc finger protein 513 |
| chr20_+_46428124 | 0.06 |
ENSRNOT00000000327
|
Foxo3
|
forkhead box O3 |
| chr17_-_90756048 | 0.05 |
ENSRNOT00000085669
|
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr3_-_120087136 | 0.05 |
ENSRNOT00000078994
|
Kcnip3
|
potassium voltage-gated channel interacting protein 3 |
| chr10_-_58693754 | 0.05 |
ENSRNOT00000071764
|
Pitpnm3
|
PITPNM family member 3 |
| chrX_+_19270181 | 0.05 |
ENSRNOT00000029397
|
Usp51
|
ubiquitin specific peptidase 51 |
| chr8_-_32328819 | 0.05 |
ENSRNOT00000074457
ENSRNOT00000070818 |
Aplp2
|
amyloid beta precursor like protein 2 |
| chr2_+_262914327 | 0.05 |
ENSRNOT00000029312
|
Negr1
|
neuronal growth regulator 1 |
| chr11_-_86890390 | 0.04 |
ENSRNOT00000046183
|
Trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr6_+_147876237 | 0.04 |
ENSRNOT00000056649
|
Tmem196
|
transmembrane protein 196 |
| chr6_-_22092346 | 0.04 |
ENSRNOT00000083128
ENSRNOT00000006830 |
Birc6
|
baculoviral IAP repeat-containing 6 |
| chr4_-_125929002 | 0.04 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr2_-_211322719 | 0.04 |
ENSRNOT00000027493
|
RGD1310209
|
similar to KIAA1324 protein |
| chr1_-_142164263 | 0.04 |
ENSRNOT00000016281
|
Man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr6_-_92527711 | 0.04 |
ENSRNOT00000007529
|
Nin
|
ninein |
| chr15_+_46784963 | 0.03 |
ENSRNOT00000015540
|
Xkr6
|
XK related 6 |
| chr15_-_56970365 | 0.03 |
ENSRNOT00000047192
|
Lrch1
|
leucine rich repeats and calponin homology domain containing 1 |
| chr19_+_54766589 | 0.03 |
ENSRNOT00000025894
|
Banp
|
Btg3 associated nuclear protein |
| chr3_+_8453609 | 0.03 |
ENSRNOT00000041921
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr7_-_12424367 | 0.03 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr18_+_27576129 | 0.03 |
ENSRNOT00000070930
|
Kdm3b
|
lysine demethylase 3B |
| chr18_-_61788859 | 0.03 |
ENSRNOT00000034075
ENSRNOT00000034069 |
Ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr15_+_39644851 | 0.03 |
ENSRNOT00000081373
|
Rcbtb1
|
RCC1 and BTB domain containing protein 1 |
| chr7_-_14189688 | 0.02 |
ENSRNOT00000037456
|
Notch3
|
notch 3 |
| chr1_+_171820423 | 0.02 |
ENSRNOT00000047831
|
Ppfibp2
|
PPFIA binding protein 2 |
| chr5_+_9279970 | 0.02 |
ENSRNOT00000067977
|
Mybl1
|
myeloblastosis oncogene-like 1 |
| chr3_+_113251778 | 0.02 |
ENSRNOT00000083005
|
Map1a
|
microtubule-associated protein 1A |
| chr10_-_104358253 | 0.02 |
ENSRNOT00000005942
|
Caskin2
|
cask-interacting protein 2 |
| chr6_-_138249382 | 0.02 |
ENSRNOT00000085678
ENSRNOT00000006912 |
Ighm
|
immunoglobulin heavy constant mu |
| chr2_+_251863069 | 0.02 |
ENSRNOT00000036282
|
Syde2
|
synapse defective Rho GTPase homolog 2 |
| chr3_+_103753238 | 0.02 |
ENSRNOT00000007144
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr1_+_140602542 | 0.02 |
ENSRNOT00000085570
|
Isg20
|
interferon stimulated exonuclease gene 20 |
| chr8_-_58647933 | 0.02 |
ENSRNOT00000038830
|
Tnfaip8l3
|
TNF alpha induced protein 8 like 3 |
| chr10_+_43538160 | 0.02 |
ENSRNOT00000089217
|
Cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr1_-_2073896 | 0.02 |
ENSRNOT00000061999
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr1_-_192025350 | 0.01 |
ENSRNOT00000071946
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chrX_+_20216587 | 0.01 |
ENSRNOT00000073114
|
AABR07037412.2
|
FYVE, RhoGEF and PH domain-containing protein 1 |
| chr1_-_188713270 | 0.01 |
ENSRNOT00000082192
ENSRNOT00000065892 |
Gprc5b
|
G protein-coupled receptor, class C, group 5, member B |
| chr12_-_6956914 | 0.01 |
ENSRNOT00000072129
ENSRNOT00000001210 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chrX_+_14498119 | 0.01 |
ENSRNOT00000051135
|
Xk
|
X-linked Kx blood group |
| chr1_+_221443896 | 0.01 |
ENSRNOT00000073942
|
Tmem262
|
transmembrane protein 262 |
| chr12_+_22026075 | 0.01 |
ENSRNOT00000029041
|
LOC100910838
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr7_+_91384187 | 0.01 |
ENSRNOT00000005828
|
Utp23
|
UTP23, small subunit processome component |
| chr1_+_192025710 | 0.01 |
ENSRNOT00000077457
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chrX_-_154722220 | 0.01 |
ENSRNOT00000089743
ENSRNOT00000087893 |
Fmr1
|
fragile X mental retardation 1 |
| chr1_+_44019059 | 0.01 |
ENSRNOT00000065733
ENSRNOT00000083425 |
Scaf8
|
SR-related CTD-associated factor 8 |
| chr1_+_192025357 | 0.01 |
ENSRNOT00000025072
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr2_-_220101791 | 0.01 |
ENSRNOT00000022905
|
Plppr5
|
phospholipid phosphatase related 5 |
| chr20_+_2194709 | 0.00 |
ENSRNOT00000001017
|
Trim15
|
tripartite motif containing 15 |
| chr6_+_147876557 | 0.00 |
ENSRNOT00000080090
|
Tmem196
|
transmembrane protein 196 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Insm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |