Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Ikzf2
Z-value: 0.42
Transcription factors associated with Ikzf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ikzf2
|
ENSRNOG00000027430 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ikzf2 | rn6_v1_chr9_-_76768770_76768806 | -0.40 | 5.0e-01 | Click! |
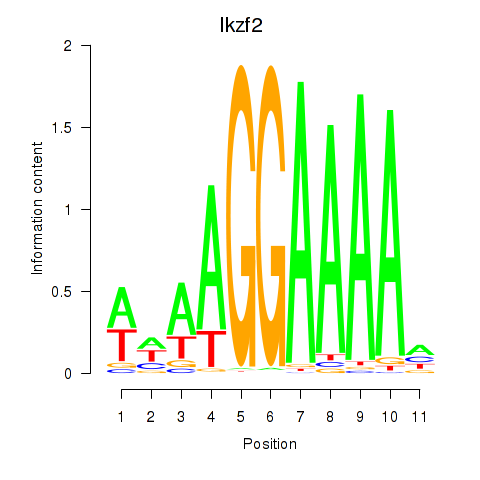
Activity profile of Ikzf2 motif
Sorted Z-values of Ikzf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_15274917 | 0.25 |
ENSRNOT00000019650
|
Pgc
|
progastricsin |
| chr2_-_208225888 | 0.14 |
ENSRNOT00000054860
|
AABR07012775.1
|
|
| chr9_+_40975836 | 0.14 |
ENSRNOT00000084470
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr15_-_33250546 | 0.12 |
ENSRNOT00000017857
|
RGD1565222
|
similar to RIKEN cDNA 4931414P19 |
| chr1_+_141767940 | 0.12 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr12_-_51965779 | 0.11 |
ENSRNOT00000056733
|
LOC100362927
|
replication protein A3-like |
| chr9_+_73433252 | 0.11 |
ENSRNOT00000092540
|
Map2
|
microtubule-associated protein 2 |
| chr1_-_131454689 | 0.11 |
ENSRNOT00000014152
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr5_+_58393233 | 0.10 |
ENSRNOT00000000142
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr4_-_117296082 | 0.10 |
ENSRNOT00000021097
|
Egr4
|
early growth response 4 |
| chr14_-_112946204 | 0.10 |
ENSRNOT00000056813
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr7_-_104541392 | 0.10 |
ENSRNOT00000078116
|
Fam49b
|
family with sequence similarity 49, member B |
| chr3_+_149790856 | 0.09 |
ENSRNOT00000075275
|
LOC100911637
|
high mobility group protein B1-like |
| chr2_+_58448917 | 0.08 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chr14_+_8080565 | 0.08 |
ENSRNOT00000092395
|
Mapk10
|
mitogen activated protein kinase 10 |
| chr10_-_38985466 | 0.08 |
ENSRNOT00000010121
|
Il13
|
interleukin 13 |
| chr13_+_98311827 | 0.07 |
ENSRNOT00000082844
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr10_-_65066612 | 0.07 |
ENSRNOT00000043448
|
Myo18a
|
myosin XVIIIa |
| chr10_-_74769637 | 0.07 |
ENSRNOT00000008889
|
LOC102555183
|
zinc finger protein OZF-like |
| chr13_-_80745347 | 0.07 |
ENSRNOT00000041908
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr12_+_25450286 | 0.06 |
ENSRNOT00000092956
|
Gtf2i
|
general transcription factor II I |
| chr6_+_99356509 | 0.06 |
ENSRNOT00000008416
|
Akap5
|
A-kinase anchoring protein 5 |
| chr9_+_52023295 | 0.06 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr2_+_236233239 | 0.06 |
ENSRNOT00000013694
|
Lef1
|
lymphoid enhancer binding factor 1 |
| chr9_-_116222374 | 0.06 |
ENSRNOT00000090111
ENSRNOT00000067900 |
Arhgap28
|
Rho GTPase activating protein 28 |
| chr11_+_84745904 | 0.06 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr7_-_120770435 | 0.06 |
ENSRNOT00000077000
|
Ddx17
|
DEAD-box helicase 17 |
| chr7_-_51515373 | 0.06 |
ENSRNOT00000080285
|
Ppp1r12a
|
protein phosphatase 1, regulatory subunit 12A |
| chrX_-_31851715 | 0.06 |
ENSRNOT00000068601
|
Vegfd
|
vascular endothelial growth factor D |
| chr10_-_16046033 | 0.06 |
ENSRNOT00000089460
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr2_-_80667481 | 0.05 |
ENSRNOT00000016784
|
Trio
|
trio Rho guanine nucleotide exchange factor |
| chr6_-_42630983 | 0.05 |
ENSRNOT00000071977
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr8_-_49158971 | 0.05 |
ENSRNOT00000020573
|
Kmt2a
|
lysine methyltransferase 2A |
| chr7_-_51515131 | 0.05 |
ENSRNOT00000006773
ENSRNOT00000041473 ENSRNOT00000050037 |
Ppp1r12a
|
protein phosphatase 1, regulatory subunit 12A |
| chr17_+_8135251 | 0.05 |
ENSRNOT00000016982
|
Trpc7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr6_-_131914028 | 0.05 |
ENSRNOT00000007602
|
Bcl11b
|
B-cell CLL/lymphoma 11B |
| chr13_+_57243877 | 0.05 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr4_-_183293999 | 0.05 |
ENSRNOT00000079695
|
Ipo8
|
importin 8 |
| chr4_-_62663200 | 0.05 |
ENSRNOT00000040969
|
Cnot4
|
CCR4-NOT transcription complex, subunit 4 |
| chr4_-_29778039 | 0.04 |
ENSRNOT00000074177
|
Sgce
|
sarcoglycan, epsilon |
| chrX_+_78042859 | 0.04 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr20_-_4542947 | 0.04 |
ENSRNOT00000086140
|
Cfb
|
complement factor B |
| chr3_-_64024205 | 0.04 |
ENSRNOT00000037015
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr13_+_90244681 | 0.04 |
ENSRNOT00000078162
|
Cd84
|
CD84 molecule |
| chr13_+_110398800 | 0.04 |
ENSRNOT00000064492
|
Lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr3_-_59688692 | 0.04 |
ENSRNOT00000078752
|
Sp3
|
Sp3 transcription factor |
| chr15_+_34493138 | 0.04 |
ENSRNOT00000089584
ENSRNOT00000027789 |
Nfatc4
|
nuclear factor of activated T-cells 4 |
| chr13_+_46169963 | 0.04 |
ENSRNOT00000005212
|
Thsd7b
|
thrombospondin type 1 domain containing 7B |
| chr1_-_260254600 | 0.04 |
ENSRNOT00000019014
|
Blnk
|
B-cell linker |
| chr10_-_21265026 | 0.04 |
ENSRNOT00000011922
|
Tenm2
|
teneurin transmembrane protein 2 |
| chr2_-_198120041 | 0.04 |
ENSRNOT00000079621
ENSRNOT00000028747 |
Plekho1
|
pleckstrin homology domain containing O1 |
| chr4_-_34194764 | 0.03 |
ENSRNOT00000045270
|
Col28a1
|
collagen type XXVIII alpha 1 chain |
| chr6_-_86713370 | 0.03 |
ENSRNOT00000005821
|
Klhl28
|
kelch-like family member 28 |
| chr9_+_71915421 | 0.03 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr6_+_86713604 | 0.03 |
ENSRNOT00000059271
|
Fam179b
|
family with sequence similarity 179, member B |
| chr4_+_180291389 | 0.03 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr1_+_266255797 | 0.03 |
ENSRNOT00000027047
|
Trim8
|
tripartite motif-containing 8 |
| chr6_-_76552559 | 0.03 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr6_+_122751016 | 0.03 |
ENSRNOT00000078050
|
Zc3h14
|
zinc finger CCCH type containing 14 |
| chr17_+_56109549 | 0.03 |
ENSRNOT00000022190
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr6_+_60566196 | 0.03 |
ENSRNOT00000006709
ENSRNOT00000075193 |
Dock4
|
dedicator of cytokinesis 4 |
| chr14_-_107160334 | 0.03 |
ENSRNOT00000012131
|
Ehbp1
|
EH domain binding protein 1 |
| chr3_+_69549673 | 0.03 |
ENSRNOT00000043974
|
Zfp804a
|
zinc finger protein 804A |
| chr4_+_22084954 | 0.03 |
ENSRNOT00000090968
|
Crot
|
carnitine O-octanoyltransferase |
| chr3_+_93920013 | 0.03 |
ENSRNOT00000083527
|
Lmo2
|
LIM domain only 2 |
| chr10_-_16045835 | 0.03 |
ENSRNOT00000064832
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr10_+_17327275 | 0.03 |
ENSRNOT00000005486
|
Ubtd2
|
ubiquitin domain containing 2 |
| chr9_+_94316717 | 0.03 |
ENSRNOT00000077005
ENSRNOT00000092534 |
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr14_+_8080275 | 0.02 |
ENSRNOT00000065965
ENSRNOT00000092542 |
Mapk10
|
mitogen activated protein kinase 10 |
| chr8_+_52127632 | 0.02 |
ENSRNOT00000079797
|
Cadm1
|
cell adhesion molecule 1 |
| chr1_+_190671696 | 0.02 |
ENSRNOT00000084934
|
AABR07005633.1
|
|
| chr18_+_30496318 | 0.02 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr18_+_30562178 | 0.02 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr6_+_28235695 | 0.02 |
ENSRNOT00000047210
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr6_-_26828972 | 0.02 |
ENSRNOT00000010932
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr10_-_92476109 | 0.02 |
ENSRNOT00000089029
|
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr2_-_41784929 | 0.02 |
ENSRNOT00000086851
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr11_-_35749464 | 0.02 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chr8_-_11887799 | 0.02 |
ENSRNOT00000072420
|
Jrkl
|
JRK-like |
| chr1_-_259106948 | 0.02 |
ENSRNOT00000074429
|
LOC100912537
|
tectonic-3-like |
| chr3_+_44025300 | 0.02 |
ENSRNOT00000006319
|
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr9_+_73334618 | 0.02 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chrX_+_84064427 | 0.02 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr13_-_89874008 | 0.02 |
ENSRNOT00000051368
|
Ptma
|
prothymosin alpha |
| chr16_+_24980723 | 0.01 |
ENSRNOT00000082142
|
Tma16
|
translation machinery associated 16 homolog |
| chr12_-_51702730 | 0.01 |
ENSRNOT00000046920
|
Ttc28
|
tetratricopeptide repeat domain 28 |
| chr3_+_113918629 | 0.01 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chrX_-_25590048 | 0.01 |
ENSRNOT00000004873
|
Mid1
|
midline 1 |
| chr10_-_87437455 | 0.01 |
ENSRNOT00000051080
|
Krt39
|
keratin 39 |
| chr20_+_27975549 | 0.01 |
ENSRNOT00000092075
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr18_+_30487264 | 0.01 |
ENSRNOT00000040125
|
Pcdhb10
|
protocadherin beta 10 |
| chr11_-_14304603 | 0.01 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr3_+_11554457 | 0.01 |
ENSRNOT00000073087
|
Fam102a
|
family with sequence similarity 102, member A |
| chr3_+_93920447 | 0.01 |
ENSRNOT00000012625
|
Lmo2
|
LIM domain only 2 |
| chr10_+_53740841 | 0.01 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr15_+_24479761 | 0.01 |
ENSRNOT00000016449
|
Ktn1
|
kinectin 1 |
| chr8_+_118744229 | 0.01 |
ENSRNOT00000028364
|
Kif9
|
kinesin family member 9 |
| chr9_+_66952720 | 0.01 |
ENSRNOT00000023678
|
Nbeal1
|
neurobeachin-like 1 |
| chr5_+_28395296 | 0.01 |
ENSRNOT00000009375
|
Tmem55a
|
transmembrane protein 55A |
| chr2_-_123851854 | 0.01 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr3_+_8873933 | 0.01 |
ENSRNOT00000030996
|
Nup188
|
nucleoporin 188 |
| chr10_-_56365084 | 0.00 |
ENSRNOT00000068013
|
Polr2a
|
RNA polymerase II subunit A |
| chr5_-_77408323 | 0.00 |
ENSRNOT00000046857
ENSRNOT00000046760 |
LOC259245
Mup4
|
alpha-2u globulin PGCL5 major urinary protein 4 |
| chr4_+_66091641 | 0.00 |
ENSRNOT00000043147
|
AABR07060287.1
|
|
| chrX_-_29648359 | 0.00 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chrX_+_78259409 | 0.00 |
ENSRNOT00000049779
|
RGD1560455
|
similar to RIKEN cDNA A630033H20 gene |
| chr18_+_61261418 | 0.00 |
ENSRNOT00000064250
|
Zfp532
|
zinc finger protein 532 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ikzf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.1 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.1 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.1 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 | 0.0 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0071899 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) regulation of cellular response to drug(GO:2001038) |
| 0.0 | 0.1 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |