Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
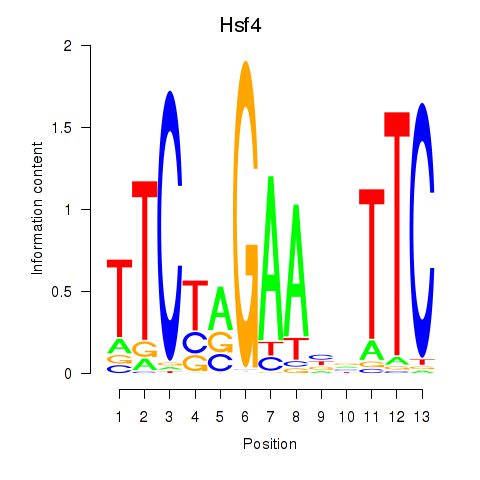
Results for Hsf4
Z-value: 0.61
Transcription factors associated with Hsf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf4
|
ENSRNOG00000015253 | heat shock transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf4 | rn6_v1_chr19_+_37229120_37229120 | 0.86 | 6.5e-02 | Click! |
Activity profile of Hsf4 motif
Sorted Z-values of Hsf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_12952906 | 0.36 |
ENSRNOT00000078193
|
LOC100360362
|
hypothetical protein LOC100360362 |
| chr5_-_153924896 | 0.34 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr1_+_83003841 | 0.31 |
ENSRNOT00000057384
|
Ceacam4
|
carcinoembryonic antigen-related cell adhesion molecule 4 |
| chr16_-_19942343 | 0.30 |
ENSRNOT00000087162
ENSRNOT00000091906 |
Bst2
|
bone marrow stromal cell antigen 2 |
| chr4_-_482645 | 0.30 |
ENSRNOT00000062073
ENSRNOT00000071713 |
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr20_-_9855443 | 0.30 |
ENSRNOT00000090275
ENSRNOT00000066266 |
Tff3
|
trefoil factor 3 |
| chr12_-_23841049 | 0.29 |
ENSRNOT00000031555
|
Hspb1
|
heat shock protein family B (small) member 1 |
| chr1_+_80135391 | 0.25 |
ENSRNOT00000021893
|
Gpr4
|
G protein-coupled receptor 4 |
| chr2_-_187668677 | 0.24 |
ENSRNOT00000056898
ENSRNOT00000092563 |
Tsacc
|
TSSK6 activating co-chaperone |
| chr8_+_55178289 | 0.24 |
ENSRNOT00000059127
|
Cryab
|
crystallin, alpha B |
| chr1_+_81643816 | 0.23 |
ENSRNOT00000027214
|
LOC103689942
|
carcinoembryonic antigen-related cell adhesion molecule 1-like |
| chr4_-_161757447 | 0.23 |
ENSRNOT00000008737
|
Fkbp4
|
FK506 binding protein 4 |
| chr3_+_154043873 | 0.22 |
ENSRNOT00000072502
ENSRNOT00000034166 |
Nnat
|
neuronatin |
| chr11_-_36479868 | 0.21 |
ENSRNOT00000075762
|
LOC100911295
|
non-histone chromosomal protein HMG-14-like |
| chr1_+_90948976 | 0.20 |
ENSRNOT00000056877
|
LOC108348111
|
succinate dehydrogenase assembly factor 1, mitochondrial |
| chr1_-_145931583 | 0.20 |
ENSRNOT00000016433
|
Cfap161
|
cilia and flagella associated protein 161 |
| chr6_-_135112775 | 0.20 |
ENSRNOT00000086310
|
LOC103692716
|
heat shock protein HSP 90-alpha |
| chr5_-_61410812 | 0.18 |
ENSRNOT00000015270
|
Igfbpl1
|
insulin-like growth factor binding protein-like 1 |
| chr17_-_44834305 | 0.17 |
ENSRNOT00000084303
|
Hist1h2bd
|
histone cluster 1, H2bd |
| chr1_+_279798187 | 0.17 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr1_-_21854763 | 0.17 |
ENSRNOT00000089196
ENSRNOT00000020528 |
Ctgf
|
connective tissue growth factor |
| chr5_-_139864393 | 0.17 |
ENSRNOT00000030016
|
Zfp69
|
zinc finger protein 69 |
| chr20_-_4921348 | 0.16 |
ENSRNOT00000082497
ENSRNOT00000041151 |
RT1-CE4
|
RT1 class I, locus CE4 |
| chr9_+_10446699 | 0.16 |
ENSRNOT00000075344
|
LOC301124
|
hypothetical LOC301124 |
| chr8_-_49301125 | 0.16 |
ENSRNOT00000091190
|
Cd3e
|
CD3e molecule |
| chr9_+_38456468 | 0.15 |
ENSRNOT00000016948
|
Bag2
|
Bcl2-associated athanogene 2 |
| chr8_+_48571323 | 0.15 |
ENSRNOT00000059776
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr20_-_46305157 | 0.15 |
ENSRNOT00000000340
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr10_-_105787803 | 0.15 |
ENSRNOT00000064671
|
Jmjd6
|
jumonji domain containing 6 |
| chr3_+_94035905 | 0.15 |
ENSRNOT00000014767
|
RGD1563222
|
similar to RIKEN cDNA A930018P22 |
| chr11_+_37798370 | 0.15 |
ENSRNOT00000002679
|
Bace2
|
beta-site APP-cleaving enzyme 2 |
| chr10_-_57436368 | 0.15 |
ENSRNOT00000056608
|
Scimp
|
SLP adaptor and CSK interacting membrane protein |
| chr11_-_61530567 | 0.15 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr3_-_7051953 | 0.15 |
ENSRNOT00000013473
|
RGD1306233
|
similar to hypothetical protein MGC29761 |
| chr1_-_222293148 | 0.14 |
ENSRNOT00000028743
|
Stip1
|
stress-induced phosphoprotein 1 |
| chr13_+_96195836 | 0.14 |
ENSRNOT00000042547
|
RGD1563812
|
similar to basic transcription factor 3 |
| chr1_-_154170409 | 0.14 |
ENSRNOT00000089014
|
Hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chr1_+_81373340 | 0.14 |
ENSRNOT00000026814
|
Zfp428
|
zinc finger protein 428 |
| chr20_+_295250 | 0.14 |
ENSRNOT00000000955
|
Clic2
|
chloride intracellular channel 2 |
| chr5_+_162351001 | 0.14 |
ENSRNOT00000065471
|
LOC691162
|
hypothetical protein LOC691162 |
| chr14_+_22706901 | 0.13 |
ENSRNOT00000088798
|
Ppial4d
|
peptidylprolyl isomerase A (cyclophilin A)-like 4D |
| chr5_+_169357517 | 0.13 |
ENSRNOT00000000079
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr1_+_89162639 | 0.13 |
ENSRNOT00000028508
|
Atp4a
|
ATPase H+/K+ transporting alpha subunit |
| chr7_-_59763219 | 0.13 |
ENSRNOT00000041210
|
RGD1564883
|
similar to 60S ribosomal protein L12 |
| chrX_+_157095937 | 0.13 |
ENSRNOT00000091792
|
Bcap31
|
B-cell receptor-associated protein 31 |
| chr6_+_99817431 | 0.12 |
ENSRNOT00000009621
|
Churc1
|
churchill domain containing 1 |
| chr1_-_164308317 | 0.12 |
ENSRNOT00000022983
|
Serpinh1
|
serpin family H member 1 |
| chr3_+_124088157 | 0.12 |
ENSRNOT00000028876
|
Smox
|
spermine oxidase |
| chr7_-_11754508 | 0.12 |
ENSRNOT00000026341
|
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr20_-_29558321 | 0.12 |
ENSRNOT00000000702
|
Anapc16
|
anaphase promoting complex subunit 16 |
| chr1_-_24190896 | 0.12 |
ENSRNOT00000016121
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr5_+_116973159 | 0.12 |
ENSRNOT00000046475
|
LOC100912024
|
uncharacterized LOC100912024 |
| chr5_+_160095427 | 0.12 |
ENSRNOT00000077609
|
AABR07050298.1
|
|
| chr6_+_98284170 | 0.12 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr8_-_87213627 | 0.11 |
ENSRNOT00000066084
|
Cox7a2
|
cytochrome c oxidase subunit 7A2 |
| chr7_-_125497691 | 0.11 |
ENSRNOT00000049445
|
AABR07058578.1
|
|
| chr5_-_127273656 | 0.11 |
ENSRNOT00000057341
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr14_-_43143973 | 0.11 |
ENSRNOT00000003248
|
Uchl1
|
ubiquitin C-terminal hydrolase L1 |
| chr15_+_32386816 | 0.11 |
ENSRNOT00000060322
|
AABR07017901.1
|
|
| chr8_+_99632803 | 0.11 |
ENSRNOT00000087190
|
Plscr1
|
phospholipid scramblase 1 |
| chr2_-_84678790 | 0.11 |
ENSRNOT00000015886
|
Cct5
|
chaperonin containing TCP1 subunit 5 |
| chr1_-_190914610 | 0.11 |
ENSRNOT00000023189
|
Cdr2
|
cerebellar degeneration-related protein 2 |
| chr10_-_13542077 | 0.10 |
ENSRNOT00000008736
|
Atp6v0c
|
ATPase H+ transporting V0 subunit C |
| chr11_+_30363280 | 0.10 |
ENSRNOT00000002885
|
Sod1
|
superoxide dismutase 1, soluble |
| chr10_+_105787935 | 0.10 |
ENSRNOT00000000266
|
Mettl23
|
methyltransferase like 23 |
| chr11_+_39482408 | 0.10 |
ENSRNOT00000075126
|
Hmgn1
|
high mobility group nucleosome binding domain 1 |
| chr3_-_111087347 | 0.10 |
ENSRNOT00000018277
|
Rhov
|
ras homolog family member V |
| chr6_-_127656603 | 0.10 |
ENSRNOT00000015516
|
Serpina11
|
serpin family A member 11 |
| chrX_-_83864150 | 0.10 |
ENSRNOT00000073734
|
Obp1f
|
odorant binding protein I f |
| chr7_+_40318490 | 0.10 |
ENSRNOT00000081374
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr3_+_111135021 | 0.10 |
ENSRNOT00000018970
|
Dll4
|
delta like canonical Notch ligand 4 |
| chr11_+_67221359 | 0.10 |
ENSRNOT00000086097
|
Casr
|
calcium-sensing receptor |
| chr5_-_155258392 | 0.10 |
ENSRNOT00000017065
|
C1qc
|
complement C1q C chain |
| chr19_+_55381565 | 0.09 |
ENSRNOT00000018923
|
Cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr3_+_149706075 | 0.09 |
ENSRNOT00000034798
|
Bpifb5
|
BPI fold containing family B, member 5 |
| chrX_-_121731543 | 0.09 |
ENSRNOT00000018788
|
Klhl13
|
kelch-like family member 13 |
| chr12_-_22021851 | 0.09 |
ENSRNOT00000039280
|
Tsc22d4
|
TSC22 domain family, member 4 |
| chrX_+_111122552 | 0.09 |
ENSRNOT00000083566
ENSRNOT00000085078 ENSRNOT00000090928 |
Cldn2
|
claudin 2 |
| chr1_+_82452469 | 0.09 |
ENSRNOT00000028026
|
Exosc5
|
exosome component 5 |
| chr10_-_107539658 | 0.09 |
ENSRNOT00000089346
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr1_+_193187972 | 0.09 |
ENSRNOT00000090172
ENSRNOT00000065342 |
Slc5a11
|
solute carrier family 5 member 11 |
| chr8_+_76426335 | 0.09 |
ENSRNOT00000085496
|
Gtf2a2
|
general transcription factor2A subunit 2 |
| chr2_+_200397967 | 0.09 |
ENSRNOT00000025821
|
Reg4
|
regenerating family member 4 |
| chr1_-_220746224 | 0.09 |
ENSRNOT00000027734
|
Banf1
|
barrier to autointegration factor 1 |
| chr1_-_175676699 | 0.09 |
ENSRNOT00000030474
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr17_-_43776460 | 0.09 |
ENSRNOT00000089055
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr9_+_81518176 | 0.09 |
ENSRNOT00000078317
ENSRNOT00000019265 ENSRNOT00000088246 ENSRNOT00000084682 |
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr2_+_88344527 | 0.09 |
ENSRNOT00000059467
|
RGD1565641
|
RGD1565641 |
| chr2_+_60461551 | 0.09 |
ENSRNOT00000024557
|
Rad1
|
RAD1 checkpoint DNA exonuclease |
| chr4_-_183426439 | 0.09 |
ENSRNOT00000083310
|
Fam60a
|
family with sequence similarity 60, member A |
| chr14_+_107767392 | 0.09 |
ENSRNOT00000012847
|
Cct4
|
chaperonin containing TCP1 subunit 4 |
| chr19_-_56677084 | 0.09 |
ENSRNOT00000024084
|
Acta1
|
actin, alpha 1, skeletal muscle |
| chr16_-_48437223 | 0.09 |
ENSRNOT00000013005
ENSRNOT00000059401 |
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr4_+_170958196 | 0.09 |
ENSRNOT00000007905
|
Pde6h
|
phosphodiesterase 6H |
| chr1_-_30917882 | 0.09 |
ENSRNOT00000015440
|
Echdc1
|
ethylmalonyl-CoA decarboxylase 1 |
| chr15_+_33632416 | 0.09 |
ENSRNOT00000068212
|
AC130940.1
|
|
| chr20_+_29558689 | 0.09 |
ENSRNOT00000085229
|
Ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr18_-_86279680 | 0.08 |
ENSRNOT00000006169
|
LOC689166
|
hypothetical protein LOC689166 |
| chr14_+_88543630 | 0.08 |
ENSRNOT00000060515
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr6_-_94980004 | 0.08 |
ENSRNOT00000006373
|
Rtn1
|
reticulon 1 |
| chr7_+_72772440 | 0.08 |
ENSRNOT00000009989
|
Mtdh
|
metadherin |
| chr2_+_27713880 | 0.08 |
ENSRNOT00000083143
|
Gcnt4
|
glucosaminyl (N-acetyl) transferase 4, core 2 (beta-1,6-N-acetylglucosaminyltransferase) |
| chr10_-_57243435 | 0.08 |
ENSRNOT00000005050
|
Chrne
|
cholinergic receptor nicotinic epsilon subunit |
| chr7_-_59882077 | 0.08 |
ENSRNOT00000068774
|
Myrfl
|
myelin regulatory factor-like |
| chr4_-_170932618 | 0.08 |
ENSRNOT00000007779
|
Arhgdib
|
Rho GDP dissociation inhibitor beta |
| chr14_+_18983853 | 0.08 |
ENSRNOT00000003836
|
Rassf6
|
Ras association domain family member 6 |
| chr20_-_5469966 | 0.08 |
ENSRNOT00000092362
|
Tapbp
|
TAP binding protein |
| chrX_-_45284341 | 0.08 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chr8_+_45561390 | 0.08 |
ENSRNOT00000047820
|
AABR07070043.1
|
|
| chr5_-_58078545 | 0.08 |
ENSRNOT00000075777
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr20_-_5037022 | 0.08 |
ENSRNOT00000091022
ENSRNOT00000060762 |
Msh5
|
mutS homolog 5 |
| chr17_+_36771639 | 0.08 |
ENSRNOT00000015147
|
Gpr50
|
G protein-coupled receptor 50 |
| chr8_+_54993859 | 0.08 |
ENSRNOT00000013093
|
Il18
|
interleukin 18 |
| chr16_+_72216326 | 0.08 |
ENSRNOT00000051363
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chrX_+_15049462 | 0.08 |
ENSRNOT00000007015
|
Ebp
|
emopamil binding protein (sterol isomerase) |
| chr10_+_3227160 | 0.07 |
ENSRNOT00000088075
|
Ntan1
|
N-terminal asparagine amidase |
| chr16_-_20807070 | 0.07 |
ENSRNOT00000072536
|
Comp
|
cartilage oligomeric matrix protein |
| chr14_-_64535170 | 0.07 |
ENSRNOT00000082338
|
Gba3
|
glucosidase, beta, acid 3 |
| chr1_-_100865894 | 0.07 |
ENSRNOT00000027580
|
Ptov1
|
prostate tumor overexpressed 1 |
| chr7_+_143060597 | 0.07 |
ENSRNOT00000087481
|
Krt7
|
keratin 7 |
| chr20_-_30327361 | 0.07 |
ENSRNOT00000000689
|
Slc29a3
|
solute carrier family 29 member 3 |
| chr20_+_4087618 | 0.07 |
ENSRNOT00000000522
ENSRNOT00000060327 ENSRNOT00000080590 |
RT1-Db1
|
RT1 class II, locus Db1 |
| chr8_+_69121682 | 0.07 |
ENSRNOT00000013461
|
Rpl4
|
ribosomal protein L4 |
| chr5_-_125345726 | 0.07 |
ENSRNOT00000011244
|
LOC688684
|
similar to 60S ribosomal protein L32 |
| chr1_-_48033343 | 0.07 |
ENSRNOT00000019531
ENSRNOT00000076422 |
Tcp1
|
t-complex 1 |
| chr1_-_170397191 | 0.07 |
ENSRNOT00000090181
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr4_-_113957774 | 0.07 |
ENSRNOT00000011921
|
Ino80b
|
INO80 complex subunit B |
| chr20_+_4966817 | 0.07 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr1_+_217039755 | 0.07 |
ENSRNOT00000091603
|
LOC102552318
|
actin-like |
| chr6_-_55647665 | 0.07 |
ENSRNOT00000007414
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr19_-_41161765 | 0.07 |
ENSRNOT00000023117
|
Hydin
|
Hydin, axonemal central pair apparatus protein |
| chr5_+_129257429 | 0.07 |
ENSRNOT00000072396
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr3_+_170994038 | 0.07 |
ENSRNOT00000081823
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chrX_+_128493614 | 0.07 |
ENSRNOT00000044240
|
Stag2
|
stromal antigen 2 |
| chr14_+_70164650 | 0.07 |
ENSRNOT00000004385
|
Qdpr
|
quinoid dihydropteridine reductase |
| chrX_-_73766885 | 0.07 |
ENSRNOT00000091325
|
Rlim
|
ring finger protein, LIM domain interacting |
| chr11_-_81379640 | 0.07 |
ENSRNOT00000002484
|
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr15_+_87886783 | 0.07 |
ENSRNOT00000065710
|
Slain1
|
SLAIN motif family, member 1 |
| chr2_-_187258140 | 0.07 |
ENSRNOT00000017735
|
Prcc
|
papillary renal cell carcinoma (translocation-associated) |
| chr11_-_38088753 | 0.06 |
ENSRNOT00000002713
|
Tmprss2
|
transmembrane protease, serine 2 |
| chr2_+_104290726 | 0.06 |
ENSRNOT00000017387
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr10_-_89199736 | 0.06 |
ENSRNOT00000027782
|
Coa3
|
cytochrome C oxidase assembly factor 3 |
| chrX_+_157150655 | 0.06 |
ENSRNOT00000090795
|
Pnck
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr5_-_164747083 | 0.06 |
ENSRNOT00000010433
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr6_-_91518996 | 0.06 |
ENSRNOT00000005835
|
Pole2
|
DNA polymerase epsilon 2, accessory subunit |
| chr1_+_165506361 | 0.06 |
ENSRNOT00000024156
|
Ucp2
|
uncoupling protein 2 |
| chr12_+_4248808 | 0.06 |
ENSRNOT00000042410
|
AABR07035089.1
|
|
| chr1_-_88780425 | 0.06 |
ENSRNOT00000074494
|
Sdhaf1
|
succinate dehydrogenase complex assembly factor 1 |
| chr16_+_6434468 | 0.06 |
ENSRNOT00000012906
|
LOC108348453
|
calsequestrin-1-like |
| chr17_-_14627937 | 0.06 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chrX_-_74847379 | 0.06 |
ENSRNOT00000067009
|
NEWGENE_1559832
|
ring finger protein, LIM domain interacting |
| chr20_-_32133431 | 0.06 |
ENSRNOT00000000443
|
Srgn
|
serglycin |
| chr4_-_85174931 | 0.06 |
ENSRNOT00000014324
ENSRNOT00000088085 |
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chrX_+_13441558 | 0.06 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr10_-_77512032 | 0.06 |
ENSRNOT00000003295
|
Pctp
|
phosphatidylcholine transfer protein |
| chr20_+_21316826 | 0.06 |
ENSRNOT00000000785
|
RGD1306739
|
similar to RIKEN cDNA 1700040L02 |
| chr1_-_102780381 | 0.06 |
ENSRNOT00000080132
|
Saa4
|
serum amyloid A4 |
| chr9_-_85626094 | 0.06 |
ENSRNOT00000020919
|
Serpine2
|
serpin family E member 2 |
| chr7_-_141185710 | 0.06 |
ENSRNOT00000085033
|
Faim2
|
Fas apoptotic inhibitory molecule 2 |
| chr8_-_109576353 | 0.06 |
ENSRNOT00000010320
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr7_-_107203897 | 0.06 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chr19_-_37952501 | 0.06 |
ENSRNOT00000026809
|
Dpep3
|
dipeptidase 3 |
| chr2_-_53413638 | 0.06 |
ENSRNOT00000021081
|
Ghr
|
growth hormone receptor |
| chr7_-_123586919 | 0.06 |
ENSRNOT00000011484
|
Ndufa6
|
NADH:ubiquinone oxidoreductase subunit A6 |
| chr10_+_43601689 | 0.06 |
ENSRNOT00000029238
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr9_+_116652530 | 0.06 |
ENSRNOT00000029210
|
L3mbtl4
|
l(3)mbt-like 4 (Drosophila) |
| chr13_+_76962504 | 0.06 |
ENSRNOT00000076581
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr4_+_52147641 | 0.06 |
ENSRNOT00000009458
|
Hyal4
|
hyaluronoglucosaminidase 4 |
| chr1_-_227175096 | 0.05 |
ENSRNOT00000054811
|
AABR07006259.1
|
|
| chr19_+_24747178 | 0.05 |
ENSRNOT00000038879
ENSRNOT00000079106 |
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr8_-_111965889 | 0.05 |
ENSRNOT00000032376
|
Bfsp2
|
beaded filament structural protein 2 |
| chr8_+_128790348 | 0.05 |
ENSRNOT00000025142
|
Slc25a38
|
solute carrier family 25, member 38 |
| chrX_-_1346181 | 0.05 |
ENSRNOT00000042736
|
LOC102555453
|
60S ribosomal protein L12-like |
| chrX_-_112159458 | 0.05 |
ENSRNOT00000087403
|
Tex13b
|
testis expressed 13B |
| chr7_+_11737293 | 0.05 |
ENSRNOT00000046059
|
Lingo3
|
leucine rich repeat and Ig domain containing 3 |
| chr17_-_27969433 | 0.05 |
ENSRNOT00000073967
|
Nrn1
|
neuritin 1 |
| chr3_+_2877293 | 0.05 |
ENSRNOT00000061855
|
Lcn5
|
lipocalin 5 |
| chr19_-_26006970 | 0.05 |
ENSRNOT00000004570
|
Gcdh
|
glutaryl-CoA dehydrogenase |
| chr12_-_31323810 | 0.05 |
ENSRNOT00000001247
|
Ran
|
RAN, member RAS oncogene family |
| chr1_+_214317682 | 0.05 |
ENSRNOT00000032904
|
Tmem80
|
transmembrane protein 80 |
| chr14_+_3058993 | 0.05 |
ENSRNOT00000002807
|
Gfi1
|
growth factor independent 1 transcriptional repressor |
| chr6_-_128690741 | 0.05 |
ENSRNOT00000035826
|
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr5_-_151768123 | 0.05 |
ENSRNOT00000079380
|
Nudc
|
nuclear distribution C, dynein complex regulator |
| chr1_-_252461461 | 0.05 |
ENSRNOT00000026093
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr19_-_26082719 | 0.05 |
ENSRNOT00000083159
|
Rnaseh2a
|
ribonuclease H2, subunit A |
| chr9_+_95161157 | 0.05 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr12_+_22153983 | 0.05 |
ENSRNOT00000080775
ENSRNOT00000001894 |
Pcolce
|
procollagen C-endopeptidase enhancer |
| chr6_+_55648021 | 0.05 |
ENSRNOT00000064822
ENSRNOT00000091488 |
Ankmy2
|
ankyrin repeat and MYND domain containing 2 |
| chr3_+_11396070 | 0.05 |
ENSRNOT00000037399
|
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr7_-_30105132 | 0.05 |
ENSRNOT00000091227
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr1_+_90729274 | 0.05 |
ENSRNOT00000047207
|
RGD1560088
|
similar to NADH:ubiquinone oxidoreductase B15 subunit |
| chr12_+_17252093 | 0.05 |
ENSRNOT00000047634
|
Zfand2a
|
zinc finger AN1-type containing 2A |
| chr2_+_25041185 | 0.05 |
ENSRNOT00000040983
|
Zbed3
|
zinc finger, BED-type containing 3 |
| chr1_-_102741581 | 0.05 |
ENSRNOT00000016254
|
LOC691143
|
similar to Serum amyloid A-3 protein precursor |
| chr1_-_86923575 | 0.05 |
ENSRNOT00000027029
|
Mrps12
|
mitochondrial ribosomal protein S12 |
| chr5_+_57028467 | 0.05 |
ENSRNOT00000010061
|
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr2_+_198312428 | 0.05 |
ENSRNOT00000028758
|
Sf3b4
|
splicing factor 3b, subunit 4 |
| chr3_+_122928964 | 0.05 |
ENSRNOT00000074724
|
LOC100365450
|
rCG26795-like |
| chr20_+_26999795 | 0.05 |
ENSRNOT00000057872
|
Mypn
|
myopalladin |
| chr1_+_214317965 | 0.05 |
ENSRNOT00000078754
|
Tmem80
|
transmembrane protein 80 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1901252 | negative regulation of dendritic cell cytokine production(GO:0002731) regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 0.3 | GO:1903544 | cellular response to interleukin-11(GO:0071348) response to butyrate(GO:1903544) |
| 0.1 | 0.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.3 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.3 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.4 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.0 | 0.1 | GO:0060816 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:0034486 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0002666 | positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.3 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0090131 | skeletal muscle thin filament assembly(GO:0030240) mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0090247 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 0.0 | 0.2 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.0 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.0 | GO:1904252 | histone H3-R17 methylation(GO:0034971) negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:2000371 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.1 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) DNA deamination(GO:0045006) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.0 | GO:1990091 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0002501 | MHC protein complex assembly(GO:0002396) MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC protein complex(GO:0002501) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.0 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.2 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.0 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.0 | 0.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0046979 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.0 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |