Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
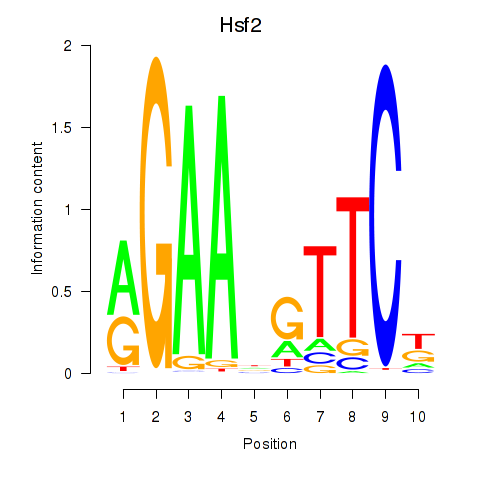
Results for Hsf2
Z-value: 0.68
Transcription factors associated with Hsf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf2
|
ENSRNOG00000000808 | heat shock transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf2 | rn6_v1_chr20_+_38935820_38935820 | -0.09 | 8.9e-01 | Click! |
Activity profile of Hsf2 motif
Sorted Z-values of Hsf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_9855443 | 0.32 |
ENSRNOT00000090275
ENSRNOT00000066266 |
Tff3
|
trefoil factor 3 |
| chr15_-_108898703 | 0.27 |
ENSRNOT00000067577
|
Zic5
|
Zic family member 5 |
| chr19_+_41482728 | 0.25 |
ENSRNOT00000022943
|
Calb2
|
calbindin 2 |
| chr4_-_482645 | 0.23 |
ENSRNOT00000062073
ENSRNOT00000071713 |
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr14_-_6679878 | 0.22 |
ENSRNOT00000075989
ENSRNOT00000067875 |
Spp1
|
secreted phosphoprotein 1 |
| chr1_-_145870912 | 0.20 |
ENSRNOT00000016289
|
Il16
|
interleukin 16 |
| chr10_+_103395511 | 0.19 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr1_+_145770135 | 0.15 |
ENSRNOT00000015320
|
Stard5
|
StAR-related lipid transfer domain containing 5 |
| chr7_+_144647587 | 0.15 |
ENSRNOT00000022398
|
Hoxc4
|
homeo box C4 |
| chr6_+_64808238 | 0.14 |
ENSRNOT00000093195
|
Nrcam
|
neuronal cell adhesion molecule |
| chr5_-_58198782 | 0.14 |
ENSRNOT00000023951
|
Ccl21
|
C-C motif chemokine ligand 21 |
| chr5_+_162031722 | 0.14 |
ENSRNOT00000020483
|
Lrrc38
|
leucine rich repeat containing 38 |
| chr9_-_65957101 | 0.13 |
ENSRNOT00000014094
|
Mpp4
|
membrane palmitoylated protein 4 |
| chr3_+_2877293 | 0.13 |
ENSRNOT00000061855
|
Lcn5
|
lipocalin 5 |
| chr8_-_48762342 | 0.13 |
ENSRNOT00000049125
|
Foxr1
|
forkhead box R1 |
| chr20_+_3149114 | 0.12 |
ENSRNOT00000084770
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr3_+_9681871 | 0.12 |
ENSRNOT00000083251
|
Prrx2
|
paired related homeobox 2 |
| chr10_+_99388130 | 0.12 |
ENSRNOT00000006238
|
Kcnj16
|
potassium voltage-gated channel subfamily J member 16 |
| chr3_-_64554953 | 0.12 |
ENSRNOT00000067452
|
LOC102553814
|
serine/arginine repetitive matrix protein 1-like |
| chr5_+_117052260 | 0.12 |
ENSRNOT00000010211
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr6_-_99843245 | 0.12 |
ENSRNOT00000080270
|
Gpx2
|
glutathione peroxidase 2 |
| chr10_+_103396155 | 0.11 |
ENSRNOT00000086924
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr5_+_154522119 | 0.11 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr1_+_82174451 | 0.11 |
ENSRNOT00000027783
|
Tmem145
|
transmembrane protein 145 |
| chr14_+_110676090 | 0.11 |
ENSRNOT00000029513
|
Fancl
|
Fanconi anemia, complementation group L |
| chr8_-_40883880 | 0.10 |
ENSRNOT00000075593
|
LOC102550797
|
disks large homolog 5-like |
| chr8_-_82339937 | 0.10 |
ENSRNOT00000080797
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr1_+_177569618 | 0.10 |
ENSRNOT00000090042
|
Tead1
|
TEA domain transcription factor 1 |
| chr7_+_120153184 | 0.10 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr2_-_195678848 | 0.10 |
ENSRNOT00000028303
ENSRNOT00000075569 |
Oaz3
|
ornithine decarboxylase antizyme 3 |
| chr16_+_20432899 | 0.10 |
ENSRNOT00000026271
|
Mpv17l2
|
MPV17 mitochondrial inner membrane protein like 2 |
| chr10_+_4957326 | 0.09 |
ENSRNOT00000003458
|
Socs1
|
suppressor of cytokine signaling 1 |
| chr10_+_84718824 | 0.09 |
ENSRNOT00000055464
|
Copz2
|
coatomer protein complex, subunit zeta 2 |
| chr7_-_123638702 | 0.09 |
ENSRNOT00000082473
ENSRNOT00000044470 ENSRNOT00000092017 |
Cyp2d1
|
cytochrome P450, family 2, subfamily d, polypeptide 1 |
| chr1_+_238222521 | 0.09 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr3_+_176439199 | 0.09 |
ENSRNOT00000068252
|
Birc7
|
baculoviral IAP repeat-containing 7 |
| chr14_-_18733391 | 0.09 |
ENSRNOT00000003745
|
Cxcl2
|
C-X-C motif chemokine ligand 2 |
| chr17_+_72429618 | 0.09 |
ENSRNOT00000026187
|
Gata3
|
GATA binding protein 3 |
| chr4_+_57823411 | 0.09 |
ENSRNOT00000030462
ENSRNOT00000088235 |
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr7_-_123088819 | 0.08 |
ENSRNOT00000056077
|
AC096601.1
|
|
| chr17_+_28504623 | 0.08 |
ENSRNOT00000021568
|
F13a1
|
coagulation factor XIII A1 chain |
| chr5_+_90818736 | 0.08 |
ENSRNOT00000093480
ENSRNOT00000009083 |
Kdm4c
|
lysine demethylase 4C |
| chr10_-_57436368 | 0.08 |
ENSRNOT00000056608
|
Scimp
|
SLP adaptor and CSK interacting membrane protein |
| chr4_+_153874852 | 0.07 |
ENSRNOT00000079744
|
Slc6a13
|
solute carrier family 6 member 13 |
| chr10_+_93305969 | 0.07 |
ENSRNOT00000008019
|
Efcab3
|
EF-hand calcium binding domain 3 |
| chr1_-_266862842 | 0.07 |
ENSRNOT00000027496
|
LOC103693430
|
up-regulated during skeletal muscle growth protein 5 |
| chr10_+_85130141 | 0.07 |
ENSRNOT00000088101
|
AABR07030425.1
|
|
| chr1_-_101095594 | 0.07 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr2_-_45077219 | 0.07 |
ENSRNOT00000014319
|
Gzmk
|
granzyme K |
| chr4_-_27726943 | 0.07 |
ENSRNOT00000068499
|
LOC500007
|
similar to EF hand calcium binding domain 1 |
| chr1_+_220137257 | 0.07 |
ENSRNOT00000026832
|
Zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr13_+_80517536 | 0.07 |
ENSRNOT00000004386
|
Myoc
|
myocilin |
| chr2_-_123396147 | 0.07 |
ENSRNOT00000079004
|
Trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr3_+_95711555 | 0.07 |
ENSRNOT00000006302
|
Pax6
|
paired box 6 |
| chr3_-_3541947 | 0.07 |
ENSRNOT00000024786
|
Nacc2
|
NACC family member 2 |
| chr14_+_115275894 | 0.07 |
ENSRNOT00000033437
|
Gpr75
|
G protein-coupled receptor 75 |
| chr10_+_103934797 | 0.07 |
ENSRNOT00000035865
|
Cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr1_+_89008117 | 0.07 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chr9_-_80166807 | 0.07 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr10_-_93679974 | 0.07 |
ENSRNOT00000009316
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr2_+_28460068 | 0.07 |
ENSRNOT00000066819
|
Foxd1
|
forkhead box D1 |
| chr17_-_80860195 | 0.07 |
ENSRNOT00000038651
|
Trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr9_-_80167033 | 0.07 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr20_+_1736377 | 0.07 |
ENSRNOT00000047035
|
Olr1734
|
olfactory receptor 1734 |
| chr1_-_102741581 | 0.07 |
ENSRNOT00000016254
|
LOC691143
|
similar to Serum amyloid A-3 protein precursor |
| chr10_-_98294522 | 0.07 |
ENSRNOT00000005489
|
Abca8
|
ATP binding cassette subfamily A member 8 |
| chr20_+_4967194 | 0.06 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr1_-_266914093 | 0.06 |
ENSRNOT00000027526
|
Calhm2
|
calcium homeostasis modulator 2 |
| chr1_+_40816107 | 0.06 |
ENSRNOT00000060767
|
Akap12
|
A-kinase anchoring protein 12 |
| chr4_+_66091641 | 0.06 |
ENSRNOT00000043147
|
AABR07060287.1
|
|
| chr1_-_173764246 | 0.06 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr1_-_220746224 | 0.06 |
ENSRNOT00000027734
|
Banf1
|
barrier to autointegration factor 1 |
| chr15_+_32386816 | 0.06 |
ENSRNOT00000060322
|
AABR07017901.1
|
|
| chr5_+_160095427 | 0.06 |
ENSRNOT00000077609
|
AABR07050298.1
|
|
| chr4_+_29535852 | 0.06 |
ENSRNOT00000087619
|
NEWGENE_621351
|
collagen, type I, alpha 2 |
| chr4_-_161757447 | 0.06 |
ENSRNOT00000008737
|
Fkbp4
|
FK506 binding protein 4 |
| chr11_-_15858281 | 0.06 |
ENSRNOT00000090586
|
AABR07033287.1
|
|
| chr16_-_73827488 | 0.06 |
ENSRNOT00000064070
|
Ank1
|
ankyrin 1 |
| chr10_+_34277993 | 0.06 |
ENSRNOT00000055872
ENSRNOT00000003343 |
Ifi47
|
interferon gamma inducible protein 47 |
| chr12_+_41200718 | 0.06 |
ENSRNOT00000038426
ENSRNOT00000048450 ENSRNOT00000067176 |
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr9_-_82154266 | 0.06 |
ENSRNOT00000024283
|
Cryba2
|
crystallin, beta A2 |
| chr8_+_99625545 | 0.06 |
ENSRNOT00000010689
ENSRNOT00000056727 |
Plscr1
|
phospholipid scramblase 1 |
| chr3_-_3660006 | 0.06 |
ENSRNOT00000045587
|
Lhx3
|
LIM homeobox 3 |
| chr4_+_78320190 | 0.06 |
ENSRNOT00000032742
ENSRNOT00000091359 |
Gimap4
|
GTPase, IMAP family member 4 |
| chr1_+_40879747 | 0.06 |
ENSRNOT00000083702
|
Akap12
|
A-kinase anchoring protein 12 |
| chr1_+_198744050 | 0.06 |
ENSRNOT00000024404
|
Itgal
|
integrin subunit alpha L |
| chr3_+_148579920 | 0.06 |
ENSRNOT00000012432
|
Hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr1_+_177495782 | 0.06 |
ENSRNOT00000021020
|
Tead1
|
TEA domain transcription factor 1 |
| chr11_+_84827062 | 0.06 |
ENSRNOT00000058006
|
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr7_+_143059764 | 0.05 |
ENSRNOT00000010660
ENSRNOT00000087370 |
Krt7
|
keratin 7 |
| chr12_-_50404550 | 0.05 |
ENSRNOT00000073072
|
Crybb1
|
crystallin, beta B1 |
| chr5_-_38923095 | 0.05 |
ENSRNOT00000009146
|
AABR07047593.1
|
|
| chr4_+_61850348 | 0.05 |
ENSRNOT00000013423
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr11_-_79703736 | 0.05 |
ENSRNOT00000044279
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr5_+_76860515 | 0.05 |
ENSRNOT00000022866
|
RGD1310951
|
similar to RIKEN cDNA E130308A19 |
| chr5_-_58078545 | 0.05 |
ENSRNOT00000075777
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr3_+_148108539 | 0.05 |
ENSRNOT00000010149
|
Rem1
|
RRAD and GEM like GTPase 1 |
| chrX_+_119390013 | 0.05 |
ENSRNOT00000074269
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr10_-_15098791 | 0.05 |
ENSRNOT00000026139
|
Chtf18
|
chromosome transmission fidelity factor 18 |
| chr4_+_102351036 | 0.05 |
ENSRNOT00000079277
|
AABR07060994.1
|
|
| chr3_+_154043873 | 0.05 |
ENSRNOT00000072502
ENSRNOT00000034166 |
Nnat
|
neuronatin |
| chr15_+_52234563 | 0.05 |
ENSRNOT00000015169
|
Reep4
|
receptor accessory protein 4 |
| chr3_+_45683993 | 0.05 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chrX_+_159158194 | 0.05 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr5_+_116973159 | 0.05 |
ENSRNOT00000046475
|
LOC100912024
|
uncharacterized LOC100912024 |
| chr18_+_55505993 | 0.05 |
ENSRNOT00000043736
|
RGD1309362
|
similar to interferon-inducible GTPase |
| chr9_+_65331290 | 0.05 |
ENSRNOT00000058667
ENSRNOT00000018014 |
Nif3l1
|
NGG1 interacting factor 3 like 1 |
| chr5_-_159602251 | 0.05 |
ENSRNOT00000011394
|
Necap2
|
NECAP endocytosis associated 2 |
| chr11_+_71938388 | 0.05 |
ENSRNOT00000043365
|
Pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr5_-_125345726 | 0.05 |
ENSRNOT00000011244
|
LOC688684
|
similar to 60S ribosomal protein L32 |
| chr10_+_73809157 | 0.05 |
ENSRNOT00000005064
|
Rnft1
|
ring finger protein, transmembrane 1 |
| chr9_+_102862890 | 0.05 |
ENSRNOT00000050494
ENSRNOT00000080129 |
Fam174a
|
family with sequence similarity 174, member A |
| chr1_-_89559960 | 0.05 |
ENSRNOT00000092133
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr18_-_29611212 | 0.05 |
ENSRNOT00000022685
|
Dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr14_-_16903242 | 0.05 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr1_+_127010588 | 0.05 |
ENSRNOT00000016939
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chr5_+_1417478 | 0.05 |
ENSRNOT00000008153
ENSRNOT00000085564 |
Jph1
|
junctophilin 1 |
| chr6_+_24163026 | 0.05 |
ENSRNOT00000061284
|
Lbh
|
limb bud and heart development |
| chr2_-_30522727 | 0.05 |
ENSRNOT00000061183
|
Gtf2h2
|
general transcription factor IIH subunit 2 |
| chr2_-_96509424 | 0.05 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chrX_+_106487870 | 0.05 |
ENSRNOT00000075521
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr19_+_38422164 | 0.05 |
ENSRNOT00000017174
|
Nqo1
|
NAD(P)H quinone dehydrogenase 1 |
| chr5_+_162351001 | 0.05 |
ENSRNOT00000065471
|
LOC691162
|
hypothetical protein LOC691162 |
| chr11_-_71743421 | 0.05 |
ENSRNOT00000002395
|
Rnf168
|
ring finger protein 168 |
| chr14_+_17195014 | 0.05 |
ENSRNOT00000031667
|
Cxcl11
|
C-X-C motif chemokine ligand 11 |
| chr10_-_13115294 | 0.05 |
ENSRNOT00000005899
|
NEWGENE_6497122
|
FLYWCH family member 2 |
| chr6_+_137063611 | 0.05 |
ENSRNOT00000017504
|
LOC691485
|
hypothetical protein LOC691485 |
| chr1_+_205706468 | 0.05 |
ENSRNOT00000089957
ENSRNOT00000023877 |
Edrf1
|
erythroid differentiation regulatory factor 1 |
| chr20_+_3156170 | 0.04 |
ENSRNOT00000082880
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr3_-_57192125 | 0.04 |
ENSRNOT00000032528
|
RGD1565767
|
similar to ribosomal protein L15 |
| chr9_+_50526811 | 0.04 |
ENSRNOT00000036990
|
RGD1305645
|
similar to RIKEN cDNA 1500015O10 |
| chr17_-_2660902 | 0.04 |
ENSRNOT00000067979
|
Mfsd14b
|
major facilitator superfamily domain containing 14B |
| chr4_+_6931495 | 0.04 |
ENSRNOT00000079770
|
Wdr86
|
WD repeat domain 86 |
| chr10_-_107539658 | 0.04 |
ENSRNOT00000089346
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr1_+_72661211 | 0.04 |
ENSRNOT00000033197
|
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr15_-_28746042 | 0.04 |
ENSRNOT00000017730
|
Sall2
|
spalt-like transcription factor 2 |
| chr8_+_131965134 | 0.04 |
ENSRNOT00000078308
|
Znf660
|
zinc finger protein 660 |
| chr4_-_115453659 | 0.04 |
ENSRNOT00000065847
|
Tex261
|
testis expressed 261 |
| chr1_-_170318935 | 0.04 |
ENSRNOT00000024119
|
Prkcdbp
|
protein kinase C, delta binding protein |
| chrX_-_111102464 | 0.04 |
ENSRNOT00000084176
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr4_-_170932618 | 0.04 |
ENSRNOT00000007779
|
Arhgdib
|
Rho GDP dissociation inhibitor beta |
| chr5_-_124642569 | 0.04 |
ENSRNOT00000010680
|
Prkaa2
|
protein kinase AMP-activated catalytic subunit alpha 2 |
| chr9_+_121831716 | 0.04 |
ENSRNOT00000056250
ENSRNOT00000056251 |
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr18_-_1946840 | 0.04 |
ENSRNOT00000041878
|
Abhd3
|
abhydrolase domain containing 3 |
| chr8_+_94243372 | 0.04 |
ENSRNOT00000012864
|
Rwdd2a
|
RWD domain containing 2A |
| chr6_-_145777767 | 0.04 |
ENSRNOT00000046799
|
Rpl32
|
ribosomal protein L32 |
| chr12_+_13090172 | 0.04 |
ENSRNOT00000092558
|
Rac1
|
ras-related C3 botulinum toxin substrate 1 |
| chr12_+_38160464 | 0.04 |
ENSRNOT00000032249
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chr1_-_199061107 | 0.04 |
ENSRNOT00000085954
|
Zfp689
|
zinc finger protein 689 |
| chr6_-_99273033 | 0.04 |
ENSRNOT00000088808
|
Tex21
|
testis expressed 21 |
| chr5_-_100647727 | 0.04 |
ENSRNOT00000067435
|
Nfib
|
nuclear factor I/B |
| chr7_+_70292565 | 0.04 |
ENSRNOT00000073237
|
Avil
|
advillin |
| chr2_-_123396386 | 0.04 |
ENSRNOT00000046700
|
Trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr1_+_101603222 | 0.04 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr14_-_18849258 | 0.04 |
ENSRNOT00000033406
|
Pf4
|
platelet factor 4 |
| chr11_-_31238026 | 0.04 |
ENSRNOT00000032366
|
RGD1562726
|
similar to Putative protein C21orf62 homolog |
| chr4_+_163162211 | 0.04 |
ENSRNOT00000082537
|
Clec1b
|
C-type lectin domain family 1, member B |
| chr6_+_110624856 | 0.04 |
ENSRNOT00000014017
|
Vash1
|
vasohibin 1 |
| chr11_-_1983513 | 0.04 |
ENSRNOT00000000907
|
Htr1f
|
5-hydroxytryptamine receptor 1F |
| chr15_-_61648267 | 0.04 |
ENSRNOT00000071805
|
Naa16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr10_-_13107771 | 0.04 |
ENSRNOT00000005879
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr11_+_47200029 | 0.04 |
ENSRNOT00000079598
|
Nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr8_+_2604962 | 0.04 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chr20_+_3148665 | 0.04 |
ENSRNOT00000086026
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr8_-_39734214 | 0.04 |
ENSRNOT00000040106
|
Slc37a2
|
solute carrier family 37 member 2 |
| chr10_-_30118873 | 0.04 |
ENSRNOT00000006063
|
Ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr16_+_21399553 | 0.04 |
ENSRNOT00000034446
|
LOC108348302
|
zinc finger protein 709-like |
| chrX_+_106523278 | 0.04 |
ENSRNOT00000070802
|
MGC109340
|
similar to Microsomal signal peptidase 23 kDa subunit (SPase 22 kDa subunit) (SPC22/23) |
| chr20_+_1749716 | 0.04 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr10_-_107539465 | 0.04 |
ENSRNOT00000004524
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr4_-_120559078 | 0.04 |
ENSRNOT00000085730
ENSRNOT00000079575 |
Kbtbd12
|
kelch repeat and BTB domain containing 12 |
| chr4_+_100465170 | 0.04 |
ENSRNOT00000019571
|
Retsat
|
retinol saturase |
| chr8_-_117211170 | 0.04 |
ENSRNOT00000071189
|
Ccdc36
|
coiled-coil domain containing 36 |
| chr1_+_115975324 | 0.04 |
ENSRNOT00000080907
|
Atp10a
|
ATPase phospholipid transporting 10A (putative) |
| chr5_+_168009393 | 0.04 |
ENSRNOT00000055506
|
Tnfrsf9
|
TNF receptor superfamily member 9 |
| chr3_+_65816569 | 0.04 |
ENSRNOT00000079672
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr15_+_51303909 | 0.03 |
ENSRNOT00000085237
ENSRNOT00000058663 |
Loxl2
|
lysyl oxidase-like 2 |
| chr7_-_141185710 | 0.03 |
ENSRNOT00000085033
|
Faim2
|
Fas apoptotic inhibitory molecule 2 |
| chr8_-_48726963 | 0.03 |
ENSRNOT00000016145
|
Trappc4
|
trafficking protein particle complex 4 |
| chr4_-_100465106 | 0.03 |
ENSRNOT00000086725
ENSRNOT00000057989 |
Elmod3
|
ELMO domain containing 3 |
| chr12_-_22006533 | 0.03 |
ENSRNOT00000059593
|
LOC100362783
|
Uncharacterized protein C7orf61 homolog |
| chr4_+_88328061 | 0.03 |
ENSRNOT00000084775
|
Vom1r87
|
vomeronasal 1 receptor 87 |
| chr1_+_221099998 | 0.03 |
ENSRNOT00000028262
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr10_-_15166457 | 0.03 |
ENSRNOT00000026676
|
Metrn
|
meteorin, glial cell differentiation regulator |
| chr1_-_116919269 | 0.03 |
ENSRNOT00000035149
|
AABR07003616.1
|
|
| chr10_+_61640015 | 0.03 |
ENSRNOT00000092714
|
Mettl16
|
methyltransferase like 16 |
| chr1_-_164308317 | 0.03 |
ENSRNOT00000022983
|
Serpinh1
|
serpin family H member 1 |
| chr7_-_11319516 | 0.03 |
ENSRNOT00000027784
|
Apba3
|
amyloid beta precursor protein binding family A member 3 |
| chr8_-_59139946 | 0.03 |
ENSRNOT00000078571
|
Cib2
|
calcium and integrin binding family member 2 |
| chr15_-_57651041 | 0.03 |
ENSRNOT00000072138
|
Spert
|
spermatid associated |
| chr18_-_74478341 | 0.03 |
ENSRNOT00000074572
|
Slc14a1
|
solute carrier family 14 member 1 |
| chr3_+_170994038 | 0.03 |
ENSRNOT00000081823
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chr3_-_35801614 | 0.03 |
ENSRNOT00000066058
|
Mmadhc
|
methylmalonic aciduria and homocystinuria, cblD type |
| chrX_+_124321551 | 0.03 |
ENSRNOT00000074486
|
LOC100910807
|
transcriptional regulator Kaiso-like |
| chr6_-_110904288 | 0.03 |
ENSRNOT00000014645
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr6_+_137323713 | 0.03 |
ENSRNOT00000029017
|
Pld4
|
phospholipase D family, member 4 |
| chr4_-_109532234 | 0.03 |
ENSRNOT00000008665
|
Reg3g
|
regenerating family member 3 gamma |
| chr1_-_62191818 | 0.03 |
ENSRNOT00000033187
|
AABR07001926.1
|
|
| chr12_-_21746236 | 0.03 |
ENSRNOT00000001869
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr2_+_150211898 | 0.03 |
ENSRNOT00000018767
|
Sucnr1
|
succinate receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.1 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.0 | 0.1 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) |
| 0.0 | 0.1 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0035566 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) negative regulation of icosanoid secretion(GO:0032304) regulation of metanephros size(GO:0035566) |
| 0.0 | 0.1 | GO:0072268 | nephrogenic mesenchyme development(GO:0072076) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.0 | GO:0043324 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 0.0 | 0.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.0 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.0 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.0 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.2 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.0 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015489 | ornithine decarboxylase inhibitor activity(GO:0008073) polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0015265 | urea channel activity(GO:0015265) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |