Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Hoxd8
Z-value: 0.26
Transcription factors associated with Hoxd8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd8
|
ENSRNOG00000042480 | homeobox D8 |
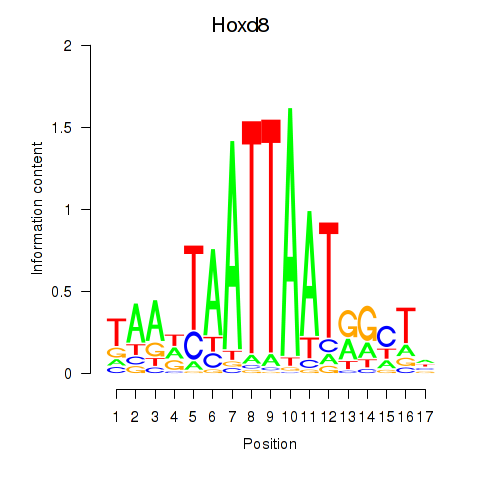
Activity profile of Hoxd8 motif
Sorted Z-values of Hoxd8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_73418607 | 0.12 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chrX_-_40086870 | 0.11 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr2_+_145174876 | 0.11 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr10_+_61685645 | 0.09 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr5_-_12199283 | 0.08 |
ENSRNOT00000007769
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr2_-_96509424 | 0.08 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr16_+_31734944 | 0.07 |
ENSRNOT00000059673
|
Palld
|
palladin, cytoskeletal associated protein |
| chr10_+_103206014 | 0.06 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr15_+_24078280 | 0.06 |
ENSRNOT00000015511
ENSRNOT00000063807 |
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr5_-_166116516 | 0.06 |
ENSRNOT00000079919
ENSRNOT00000080888 |
Kif1b
|
kinesin family member 1B |
| chr1_-_90149991 | 0.06 |
ENSRNOT00000076987
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr16_+_84465656 | 0.06 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr9_+_71915421 | 0.06 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr2_-_33025271 | 0.06 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr10_+_764421 | 0.05 |
ENSRNOT00000084608
ENSRNOT00000087567 |
Myh11
|
myosin heavy chain 11 |
| chr3_+_63379031 | 0.05 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr12_+_25435567 | 0.05 |
ENSRNOT00000092982
|
Gtf2i
|
general transcription factor II I |
| chr1_-_164977633 | 0.05 |
ENSRNOT00000029629
|
Rnf169
|
ring finger protein 169 |
| chr6_-_98666007 | 0.05 |
ENSRNOT00000082695
|
AABR07064867.1
|
|
| chr11_+_65743892 | 0.05 |
ENSRNOT00000085226
|
AC106292.2
|
|
| chr13_+_105684420 | 0.05 |
ENSRNOT00000040543
|
Gpatch2
|
G patch domain containing 2 |
| chr15_+_39638510 | 0.05 |
ENSRNOT00000037800
|
Rcbtb1
|
RCC1 and BTB domain containing protein 1 |
| chr7_+_42304534 | 0.05 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr10_-_87286387 | 0.05 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr1_-_128287151 | 0.05 |
ENSRNOT00000084946
ENSRNOT00000089723 |
Mef2a
|
myocyte enhancer factor 2a |
| chr4_-_66955732 | 0.05 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr18_-_43945273 | 0.05 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr2_-_200003443 | 0.05 |
ENSRNOT00000024900
ENSRNOT00000088041 |
Pde4dip
|
phosphodiesterase 4D interacting protein |
| chr4_-_151428894 | 0.05 |
ENSRNOT00000010556
|
Adipor2
|
adiponectin receptor 2 |
| chr14_-_82171480 | 0.04 |
ENSRNOT00000021952
|
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr2_-_231521052 | 0.04 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr17_-_89923423 | 0.04 |
ENSRNOT00000076964
|
Acbd5
|
acyl-CoA binding domain containing 5 |
| chr5_+_116420690 | 0.04 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr2_+_127525285 | 0.04 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr19_-_22194740 | 0.04 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr2_-_113345577 | 0.04 |
ENSRNOT00000034096
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr1_+_248132090 | 0.04 |
ENSRNOT00000022056
|
Il33
|
interleukin 33 |
| chr3_+_48106099 | 0.04 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr13_-_100450209 | 0.04 |
ENSRNOT00000090712
|
Lbr
|
lamin B receptor |
| chr15_-_93765498 | 0.04 |
ENSRNOT00000093297
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr13_-_94859390 | 0.04 |
ENSRNOT00000005467
ENSRNOT00000079763 |
AABR07021840.1
|
|
| chr17_+_54298409 | 0.04 |
ENSRNOT00000084919
|
Arhgap12
|
Rho GTPase activating protein 12 |
| chr13_+_43818010 | 0.04 |
ENSRNOT00000005532
|
LOC100909664
|
centrosomal protein of 170 kDa-like |
| chr9_+_52687868 | 0.04 |
ENSRNOT00000083864
|
Wdr75
|
WD repeat domain 75 |
| chr13_-_83457888 | 0.03 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr7_-_73883812 | 0.03 |
ENSRNOT00000065722
|
Stk3
|
serine/threonine kinase 3 |
| chr11_-_37253776 | 0.03 |
ENSRNOT00000002215
|
Dscam
|
DS cell adhesion molecule |
| chr17_-_79085076 | 0.03 |
ENSRNOT00000057851
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr18_+_63599425 | 0.03 |
ENSRNOT00000023145
|
Cep192
|
centrosomal protein 192 |
| chr9_+_73378057 | 0.03 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr11_-_83300409 | 0.03 |
ENSRNOT00000029255
|
Vps8
|
VPS8 CORVET complex subunit |
| chr7_-_15073052 | 0.03 |
ENSRNOT00000037708
|
Zfp799
|
zinc finger protein 799 |
| chr16_-_9658484 | 0.03 |
ENSRNOT00000065216
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chrX_+_123770337 | 0.02 |
ENSRNOT00000092554
|
Nkap
|
NFKB activating protein |
| chr7_-_70319346 | 0.02 |
ENSRNOT00000071543
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr8_+_133210473 | 0.02 |
ENSRNOT00000082774
|
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr3_+_161121697 | 0.02 |
ENSRNOT00000036020
|
Wfdc10a
|
WAP four-disulfide core domain 10A |
| chr5_-_100977902 | 0.02 |
ENSRNOT00000077429
|
Zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr10_+_70136855 | 0.02 |
ENSRNOT00000084076
|
Lig3
|
DNA ligase 3 |
| chr12_+_47590154 | 0.02 |
ENSRNOT00000045946
|
Git2
|
GIT ArfGAP 2 |
| chr19_-_37796089 | 0.02 |
ENSRNOT00000024891
|
Ranbp10
|
RAN binding protein 10 |
| chr18_+_47456327 | 0.02 |
ENSRNOT00000020030
|
Srfbp1
|
serum response factor binding protein 1 |
| chr12_-_2245324 | 0.02 |
ENSRNOT00000001332
ENSRNOT00000001333 |
Fcer2
|
Fc fragment of IgE receptor II |
| chr3_+_113918629 | 0.02 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr7_+_70987861 | 0.02 |
ENSRNOT00000072906
|
Nemp1
|
nuclear envelope integral membrane protein 1 |
| chr8_-_21453190 | 0.02 |
ENSRNOT00000078192
|
Zfp26
|
zinc finger protein 26 |
| chr19_-_29968424 | 0.02 |
ENSRNOT00000024981
|
Inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr2_-_166682325 | 0.02 |
ENSRNOT00000091198
ENSRNOT00000012422 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr9_+_10941613 | 0.02 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr10_+_67810655 | 0.02 |
ENSRNOT00000064285
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr13_+_27465930 | 0.02 |
ENSRNOT00000003314
|
Serpinb10
|
serpin family B member 10 |
| chr5_+_147185474 | 0.02 |
ENSRNOT00000000134
|
Ak2
|
adenylate kinase 2 |
| chr17_+_36691875 | 0.02 |
ENSRNOT00000074314
|
LOC688583
|
similar to High mobility group protein 4 (HMG-4) (High mobility group protein 2a) (HMG-2a) |
| chrX_-_82743753 | 0.01 |
ENSRNOT00000003512
|
Rps6ka6
|
ribosomal protein S6 kinase A6 |
| chr18_-_85980833 | 0.01 |
ENSRNOT00000058179
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr1_+_15642153 | 0.01 |
ENSRNOT00000079845
|
Map7
|
microtubule-associated protein 7 |
| chr1_+_273854248 | 0.01 |
ENSRNOT00000044827
ENSRNOT00000017600 ENSRNOT00000086496 |
Add3
|
adducin 3 |
| chr3_-_37854561 | 0.01 |
ENSRNOT00000076095
|
Neb
|
nebulin |
| chr10_+_67810810 | 0.01 |
ENSRNOT00000079156
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr6_-_38007162 | 0.01 |
ENSRNOT00000044583
|
AABR07063632.1
|
|
| chr3_-_104018861 | 0.01 |
ENSRNOT00000008387
|
Chrm5
|
cholinergic receptor, muscarinic 5 |
| chr4_+_68635581 | 0.01 |
ENSRNOT00000016217
|
Ssbp1
|
single stranded DNA binding protein 1 |
| chr5_-_56567320 | 0.01 |
ENSRNOT00000066081
|
Topors
|
TOP1 binding arginine/serine rich protein |
| chr1_+_107262659 | 0.01 |
ENSRNOT00000022499
|
Gas2
|
growth arrest-specific 2 |
| chr13_+_96303703 | 0.01 |
ENSRNOT00000084718
ENSRNOT00000029723 |
Efcab2
|
EF-hand calcium binding domain 2 |
| chr16_+_70081035 | 0.01 |
ENSRNOT00000016098
|
LOC100912059
|
ubiquitin carboxyl-terminal hydrolase 12-like |
| chr2_+_220432037 | 0.01 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr9_-_11108741 | 0.01 |
ENSRNOT00000072357
|
Ccdc94
|
coiled-coil domain containing 94 |
| chr4_+_65112944 | 0.01 |
ENSRNOT00000083672
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr8_-_126390801 | 0.01 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chr12_+_38377034 | 0.01 |
ENSRNOT00000088002
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr12_-_46493203 | 0.01 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr1_+_88955440 | 0.01 |
ENSRNOT00000091101
|
Prodh2
|
proline dehydrogenase 2 |
| chr17_-_27279260 | 0.01 |
ENSRNOT00000018508
|
Snrnp48
|
small nuclear ribonucleoprotein U11/U12 subunit 48 |
| chr8_-_118436894 | 0.01 |
ENSRNOT00000073395
|
LOC100911807
|
ATR-interacting protein-like |
| chr12_-_10967326 | 0.01 |
ENSRNOT00000042095
|
LOC100912059
|
ubiquitin carboxyl-terminal hydrolase 12-like |
| chr14_+_6221085 | 0.01 |
ENSRNOT00000068215
|
AABR07014259.1
|
|
| chrX_+_144994139 | 0.01 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr11_-_60547201 | 0.01 |
ENSRNOT00000093151
|
Btla
|
B and T lymphocyte associated |
| chr6_+_18880737 | 0.01 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr1_+_213511874 | 0.01 |
ENSRNOT00000078080
ENSRNOT00000016883 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr2_+_30685840 | 0.00 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chr3_-_45169118 | 0.00 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr3_-_22459257 | 0.00 |
ENSRNOT00000091747
|
Dennd1a
|
DENN domain containing 1A |
| chr2_+_86996497 | 0.00 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr14_+_81858737 | 0.00 |
ENSRNOT00000029784
ENSRNOT00000058068 ENSRNOT00000058067 |
Poln
|
DNA polymerase nu |
| chr8_+_1459526 | 0.00 |
ENSRNOT00000034503
|
Kbtbd3
|
kelch repeat and BTB domain containing 3 |
| chr5_+_103251986 | 0.00 |
ENSRNOT00000008757
|
Cntln
|
centlein |
| chr18_+_17043903 | 0.00 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chrX_-_76925195 | 0.00 |
ENSRNOT00000087977
|
Atrx
|
ATRX, chromatin remodeler |
| chr13_-_90405591 | 0.00 |
ENSRNOT00000006849
|
Vangl2
|
VANGL planar cell polarity protein 2 |
| chrX_+_54390733 | 0.00 |
ENSRNOT00000004977
|
RGD1565785
|
similar to chromosome X open reading frame 21 |
| chr13_-_67206688 | 0.00 |
ENSRNOT00000003630
ENSRNOT00000090693 |
Pla2g4a
|
phospholipase A2 group IVA |
| chr19_+_6046665 | 0.00 |
ENSRNOT00000084126
|
Cdh8
|
cadherin 8 |
| chr8_+_100260049 | 0.00 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.0 | GO:0070666 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.0 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |