Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Hoxd12
Z-value: 0.25
Transcription factors associated with Hoxd12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
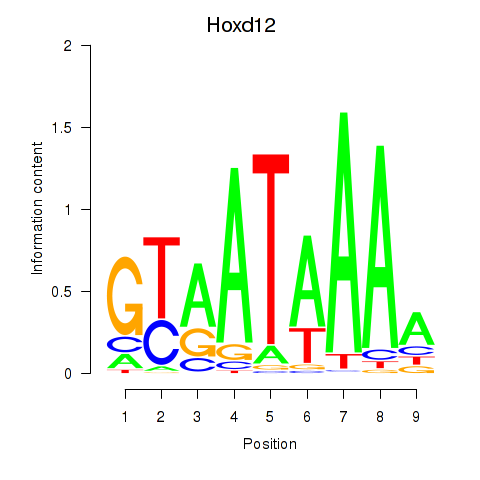
Hoxd12
|
ENSRNOG00000001587 | homeo box D12 |
Activity profile of Hoxd12 motif
Sorted Z-values of Hoxd12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_146784915 | 0.12 |
ENSRNOT00000008362
|
Sp8
|
Sp8 transcription factor |
| chr3_+_59981959 | 0.11 |
ENSRNOT00000030460
|
Sp9
|
Sp9 transcription factor |
| chr2_-_173087648 | 0.07 |
ENSRNOT00000091079
|
Iqcj
|
IQ motif containing J |
| chr10_+_77537340 | 0.07 |
ENSRNOT00000003297
|
Tmem100
|
transmembrane protein 100 |
| chr2_-_31753528 | 0.06 |
ENSRNOT00000075057
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr1_+_282134981 | 0.06 |
ENSRNOT00000036203
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr9_-_52238564 | 0.05 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr19_-_19315357 | 0.05 |
ENSRNOT00000018888
|
Cyld
|
CYLD lysine 63 deubiquitinase |
| chr13_+_57243877 | 0.05 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr7_+_28654733 | 0.04 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr9_+_19451630 | 0.04 |
ENSRNOT00000065048
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr15_-_43542939 | 0.04 |
ENSRNOT00000012996
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr13_+_90533365 | 0.04 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr14_+_12218553 | 0.03 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr2_+_8732932 | 0.03 |
ENSRNOT00000072849
|
Arrdc3
|
arrestin domain containing 3 |
| chr6_+_83421882 | 0.03 |
ENSRNOT00000084258
|
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr13_+_44615553 | 0.03 |
ENSRNOT00000082439
ENSRNOT00000005289 |
Rab3gap1
|
RAB3 GTPase activating protein catalytic subunit 1 |
| chr18_+_30880020 | 0.03 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr19_-_22194740 | 0.03 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr8_-_84632817 | 0.02 |
ENSRNOT00000076942
|
Mlip
|
muscular LMNA-interacting protein |
| chr3_+_14889510 | 0.02 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr20_+_5049496 | 0.02 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr9_+_73529612 | 0.02 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr1_-_222242644 | 0.02 |
ENSRNOT00000050891
|
Vegfb
|
vascular endothelial growth factor B |
| chrX_-_136150209 | 0.02 |
ENSRNOT00000048498
|
Enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr9_+_118586179 | 0.02 |
ENSRNOT00000022351
|
Dlgap1
|
DLG associated protein 1 |
| chr10_-_37311625 | 0.02 |
ENSRNOT00000043343
|
Jade2
|
jade family PHD finger 2 |
| chr3_+_151126591 | 0.02 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr9_-_30844199 | 0.02 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr1_-_163554839 | 0.02 |
ENSRNOT00000081509
|
Emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr8_+_103938520 | 0.02 |
ENSRNOT00000067835
|
Gk5
|
glycerol kinase 5 (putative) |
| chr4_-_179512471 | 0.02 |
ENSRNOT00000012588
|
Kras
|
KRAS proto-oncogene, GTPase |
| chr8_-_22086095 | 0.02 |
ENSRNOT00000068598
|
Zglp1
|
zinc finger, GATA-like protein 1 |
| chr14_-_16903242 | 0.02 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr3_-_67787990 | 0.02 |
ENSRNOT00000064851
|
Nckap1
|
NCK-associated protein 1 |
| chr8_-_133128290 | 0.02 |
ENSRNOT00000008783
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr2_+_52431601 | 0.02 |
ENSRNOT00000023554
|
Hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chr15_-_48284548 | 0.01 |
ENSRNOT00000038336
|
Hmbox1
|
homeobox containing 1 |
| chr7_-_59547174 | 0.01 |
ENSRNOT00000085836
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr2_+_185846232 | 0.01 |
ENSRNOT00000023418
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr13_-_107886476 | 0.01 |
ENSRNOT00000077282
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr3_-_90751055 | 0.01 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr5_-_50221095 | 0.01 |
ENSRNOT00000012164
|
RGD1563056
|
similar to hypothetical protein |
| chr7_-_134722215 | 0.01 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr7_+_140829076 | 0.01 |
ENSRNOT00000086179
|
Spats2
|
spermatogenesis associated, serine-rich 2 |
| chr10_+_70262361 | 0.01 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr2_+_226563050 | 0.01 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr15_-_55209342 | 0.01 |
ENSRNOT00000021752
|
Rb1
|
RB transcriptional corepressor 1 |
| chr3_+_48096954 | 0.01 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr5_+_4373626 | 0.01 |
ENSRNOT00000064946
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr8_+_106816152 | 0.01 |
ENSRNOT00000047613
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr17_+_78876111 | 0.01 |
ENSRNOT00000057895
|
Rpl34-ps1
|
ribosomal protein L34, pseudogene 1 |
| chr2_+_184230459 | 0.01 |
ENSRNOT00000074187
|
AABR07012054.1
|
|
| chr7_-_93280009 | 0.01 |
ENSRNOT00000064877
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr5_+_64566804 | 0.01 |
ENSRNOT00000073192
|
LOC103690035
|
TGF-beta receptor type-1-like |
| chr8_+_107229832 | 0.01 |
ENSRNOT00000045821
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr1_+_255579474 | 0.01 |
ENSRNOT00000024465
|
Btaf1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr18_+_14756684 | 0.01 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr7_+_123168811 | 0.01 |
ENSRNOT00000007091
|
Csdc2
|
cold shock domain containing C2 |
| chr4_-_176679815 | 0.01 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr1_+_78546012 | 0.01 |
ENSRNOT00000058172
|
Gng5
|
G protein subunit gamma 5 |
| chr10_-_87248572 | 0.01 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr16_+_22361998 | 0.01 |
ENSRNOT00000016193
|
Slc18a1
|
solute carrier family 18 member A1 |
| chr8_-_6235967 | 0.00 |
ENSRNOT00000068290
|
LOC654482
|
hypothetical protein LOC654482 |
| chr9_+_66058047 | 0.00 |
ENSRNOT00000071819
|
Cdk15
|
cyclin-dependent kinase 15 |
| chr8_+_59507990 | 0.00 |
ENSRNOT00000018003
|
Hykk
|
hydroxylysine kinase |
| chr5_-_154965027 | 0.00 |
ENSRNOT00000080881
ENSRNOT00000055992 |
Kdm1a
|
lysine demethylase 1A |
| chr3_-_105663457 | 0.00 |
ENSRNOT00000067646
|
Zfp770
|
zinc finger protein 770 |
| chr1_+_143036218 | 0.00 |
ENSRNOT00000025797
|
Pde8a
|
phosphodiesterase 8A |
| chr5_-_115387377 | 0.00 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr1_-_93949187 | 0.00 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr13_+_77485113 | 0.00 |
ENSRNOT00000080254
|
Tnr
|
tenascin R |
| chr2_+_188745503 | 0.00 |
ENSRNOT00000056652
|
Shc1
|
SHC adaptor protein 1 |
| chr1_+_99616447 | 0.00 |
ENSRNOT00000029197
|
Klk14
|
kallikrein related-peptidase 14 |
| chr14_+_104480662 | 0.00 |
ENSRNOT00000078710
|
Rab1a
|
RAB1A, member RAS oncogene family |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |