Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
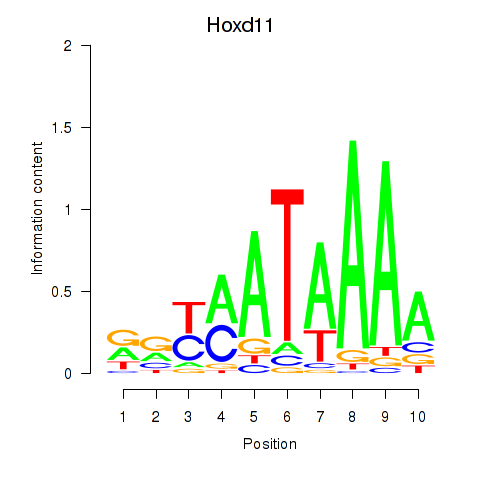
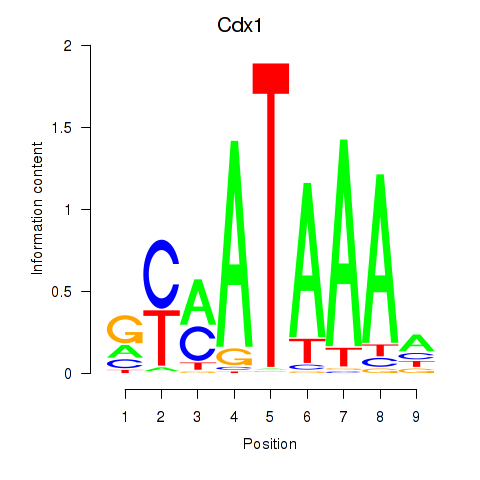
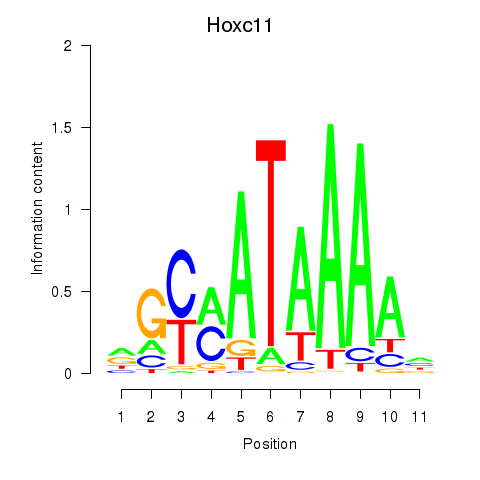
Results for Hoxd11_Cdx1_Hoxc11
Z-value: 0.21
Transcription factors associated with Hoxd11_Cdx1_Hoxc11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd11
|
ENSRNOG00000051472 | homeobox D11 |
|
Cdx1
|
ENSRNOG00000060334 | caudal type homeo box 1 |
|
Hoxc11
|
ENSRNOG00000016141 | homeobox C11 |
Activity profile of Hoxd11_Cdx1_Hoxc11 motif
Sorted Z-values of Hoxd11_Cdx1_Hoxc11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_52238564 | 0.20 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr2_-_31753528 | 0.16 |
ENSRNOT00000075057
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr19_-_22194740 | 0.14 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr6_+_146784915 | 0.12 |
ENSRNOT00000008362
|
Sp8
|
Sp8 transcription factor |
| chr19_-_19315357 | 0.10 |
ENSRNOT00000018888
|
Cyld
|
CYLD lysine 63 deubiquitinase |
| chr14_+_12218553 | 0.09 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr13_+_57243877 | 0.08 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr2_+_143895982 | 0.08 |
ENSRNOT00000080026
|
Supt20h
|
SPT20 homolog, SAGA complex component |
| chr1_+_8310577 | 0.07 |
ENSRNOT00000015131
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr15_-_48284548 | 0.06 |
ENSRNOT00000038336
|
Hmbox1
|
homeobox containing 1 |
| chr9_+_118586179 | 0.06 |
ENSRNOT00000022351
|
Dlgap1
|
DLG associated protein 1 |
| chr13_+_90533365 | 0.06 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr3_-_105663457 | 0.06 |
ENSRNOT00000067646
|
Zfp770
|
zinc finger protein 770 |
| chr16_+_22361998 | 0.05 |
ENSRNOT00000016193
|
Slc18a1
|
solute carrier family 18 member A1 |
| chr9_+_73529612 | 0.05 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr15_-_43542939 | 0.05 |
ENSRNOT00000012996
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr7_+_64769089 | 0.05 |
ENSRNOT00000088861
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr2_-_96509424 | 0.05 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr7_-_140437467 | 0.05 |
ENSRNOT00000087181
|
Fkbp11
|
FK506 binding protein 11 |
| chr5_-_154965027 | 0.04 |
ENSRNOT00000080881
ENSRNOT00000055992 |
Kdm1a
|
lysine demethylase 1A |
| chr11_-_45124423 | 0.04 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr6_-_91250138 | 0.04 |
ENSRNOT00000052408
|
LOC100911256
|
ninein-like |
| chr1_-_163554839 | 0.04 |
ENSRNOT00000081509
|
Emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr4_-_155740193 | 0.04 |
ENSRNOT00000043229
|
LOC102550396
|
LRRGT00188 |
| chr2_-_231409988 | 0.04 |
ENSRNOT00000080637
ENSRNOT00000037255 ENSRNOT00000074002 |
Ank2
|
ankyrin 2 |
| chr10_+_77537340 | 0.04 |
ENSRNOT00000003297
|
Tmem100
|
transmembrane protein 100 |
| chr2_-_173087648 | 0.04 |
ENSRNOT00000091079
|
Iqcj
|
IQ motif containing J |
| chr10_+_71546977 | 0.04 |
ENSRNOT00000041140
|
Acaca
|
acetyl-CoA carboxylase alpha |
| chr7_+_122433099 | 0.04 |
ENSRNOT00000055447
|
Tmsb10
|
thymosin, beta 10 |
| chr3_+_47677720 | 0.04 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr5_+_167141875 | 0.04 |
ENSRNOT00000089314
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr4_-_108717309 | 0.03 |
ENSRNOT00000085062
|
AABR07061178.1
|
|
| chr4_+_5644260 | 0.03 |
ENSRNOT00000041876
|
Actr3b
|
ARP3 actin related protein 3 homolog B |
| chr14_-_3288017 | 0.03 |
ENSRNOT00000080452
|
LOC689986
|
hypothetical protein LOC689986 |
| chr17_-_51912496 | 0.03 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr10_-_58771908 | 0.03 |
ENSRNOT00000018808
|
RGD1304728
|
similar to 4933427D14Rik protein |
| chr15_-_106678918 | 0.03 |
ENSRNOT00000034723
|
Stk24
|
serine/threonine kinase 24 |
| chr8_-_49158971 | 0.03 |
ENSRNOT00000020573
|
Kmt2a
|
lysine methyltransferase 2A |
| chr17_-_18590536 | 0.03 |
ENSRNOT00000078992
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr2_+_22910236 | 0.03 |
ENSRNOT00000078266
|
Homer1
|
homer scaffolding protein 1 |
| chr3_+_151126591 | 0.03 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr1_-_48891130 | 0.03 |
ENSRNOT00000083884
|
AC135026.1
|
|
| chrX_-_26376467 | 0.03 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr8_+_118066988 | 0.03 |
ENSRNOT00000056161
|
Map4
|
microtubule-associated protein 4 |
| chr15_-_55209342 | 0.03 |
ENSRNOT00000021752
|
Rb1
|
RB transcriptional corepressor 1 |
| chr2_-_231409496 | 0.03 |
ENSRNOT00000055615
ENSRNOT00000015386 ENSRNOT00000068415 |
Ank2
|
ankyrin 2 |
| chr14_+_39663421 | 0.03 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr9_-_119818310 | 0.03 |
ENSRNOT00000019359
|
Smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr7_-_118332577 | 0.02 |
ENSRNOT00000090370
|
Rbfox2
|
RNA binding protein, fox-1 homolog 2 |
| chr3_-_7796385 | 0.02 |
ENSRNOT00000082489
|
Ntng2
|
netrin G2 |
| chr1_-_200548317 | 0.02 |
ENSRNOT00000088610
|
AABR07005806.1
|
|
| chr10_-_88060561 | 0.02 |
ENSRNOT00000019133
|
Krt19
|
keratin 19 |
| chr1_+_44311513 | 0.02 |
ENSRNOT00000065386
|
Tiam2
|
T-cell lymphoma invasion and metastasis 2 |
| chr3_+_54253949 | 0.02 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr7_+_34533543 | 0.02 |
ENSRNOT00000007412
ENSRNOT00000084185 |
Ntn4
|
netrin 4 |
| chr7_-_52404774 | 0.02 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chrX_+_78042859 | 0.02 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr15_+_56666012 | 0.02 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr2_+_206064179 | 0.02 |
ENSRNOT00000025953
|
Syt6
|
synaptotagmin 6 |
| chr12_+_51960262 | 0.02 |
ENSRNOT00000084214
|
Ep400
|
E1A binding protein p400 |
| chr6_+_83421882 | 0.02 |
ENSRNOT00000084258
|
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr7_+_130042947 | 0.02 |
ENSRNOT00000089707
|
Panx2
|
pannexin 2 |
| chr8_-_63750531 | 0.02 |
ENSRNOT00000009496
|
Neo1
|
neogenin 1 |
| chr3_+_48096954 | 0.02 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr7_-_122329443 | 0.02 |
ENSRNOT00000032232
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chrX_-_10031167 | 0.02 |
ENSRNOT00000060988
|
Gpr34
|
G protein-coupled receptor 34 |
| chr6_+_107603580 | 0.02 |
ENSRNOT00000036430
|
Dnal1
|
dynein, axonemal, light chain 1 |
| chr7_+_142575672 | 0.02 |
ENSRNOT00000080923
ENSRNOT00000008160 |
Scn8a
|
sodium voltage-gated channel alpha subunit 8 |
| chr5_-_50221095 | 0.02 |
ENSRNOT00000012164
|
RGD1563056
|
similar to hypothetical protein |
| chr4_-_178441547 | 0.02 |
ENSRNOT00000087434
ENSRNOT00000055542 |
Sox5
|
SRY box 5 |
| chr3_+_43255567 | 0.02 |
ENSRNOT00000044419
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr10_-_37311625 | 0.02 |
ENSRNOT00000043343
|
Jade2
|
jade family PHD finger 2 |
| chr5_+_25725683 | 0.02 |
ENSRNOT00000087602
|
LOC679087
|
similar to swan |
| chr6_-_8344574 | 0.02 |
ENSRNOT00000009660
|
Prepl
|
prolyl endopeptidase-like |
| chr6_+_25076147 | 0.02 |
ENSRNOT00000010419
|
Ehd3
|
EH-domain containing 3 |
| chr16_-_32421005 | 0.01 |
ENSRNOT00000082712
|
Nek1
|
NIMA-related kinase 1 |
| chr13_-_37697569 | 0.01 |
ENSRNOT00000045263
|
LOC680288
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr13_-_72744295 | 0.01 |
ENSRNOT00000093366
ENSRNOT00000077157 |
Ier5
|
immediate early response 5 |
| chr13_+_77485113 | 0.01 |
ENSRNOT00000080254
|
Tnr
|
tenascin R |
| chr3_-_7795758 | 0.01 |
ENSRNOT00000045919
|
Ntng2
|
netrin G2 |
| chr4_+_158243086 | 0.01 |
ENSRNOT00000032112
|
Ano2
|
anoctamin 2 |
| chr5_+_25253010 | 0.01 |
ENSRNOT00000061333
|
Rbm12b
|
RNA binding motif protein 12B |
| chrX_+_71528988 | 0.01 |
ENSRNOT00000075963
|
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr7_-_2961873 | 0.01 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chr8_+_82037977 | 0.01 |
ENSRNOT00000082288
|
Myo5a
|
myosin VA |
| chr11_-_46369577 | 0.01 |
ENSRNOT00000049859
ENSRNOT00000047817 ENSRNOT00000002220 ENSRNOT00000002223 ENSRNOT00000063864 ENSRNOT00000070871 ENSRNOT00000072140 |
Abi3bp
|
ABI family member 3 binding protein |
| chr9_+_16737642 | 0.01 |
ENSRNOT00000061430
|
Srf
|
serum response factor |
| chr6_-_42002819 | 0.01 |
ENSRNOT00000032417
|
Greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr10_-_87248572 | 0.01 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr7_-_59547174 | 0.01 |
ENSRNOT00000085836
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr19_+_9895121 | 0.01 |
ENSRNOT00000033953
|
Prss54
|
protease, serine, 54 |
| chr5_-_6186329 | 0.01 |
ENSRNOT00000012610
|
Sulf1
|
sulfatase 1 |
| chr3_-_121882726 | 0.01 |
ENSRNOT00000006308
|
Il1b
|
interleukin 1 beta |
| chr8_+_103938520 | 0.01 |
ENSRNOT00000067835
|
Gk5
|
glycerol kinase 5 (putative) |
| chr1_+_198682230 | 0.01 |
ENSRNOT00000023995
|
Znf48
|
zinc finger protein 48 |
| chr1_+_282134981 | 0.01 |
ENSRNOT00000036203
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr4_+_6559545 | 0.01 |
ENSRNOT00000067183
|
Prkag2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr2_+_41467064 | 0.01 |
ENSRNOT00000073231
|
AABR07008066.2
|
|
| chr7_-_134722215 | 0.01 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr5_+_71742911 | 0.01 |
ENSRNOT00000047225
|
Zfp462
|
zinc finger protein 462 |
| chr7_+_80750725 | 0.01 |
ENSRNOT00000079962
|
Oxr1
|
oxidation resistance 1 |
| chr3_-_60166013 | 0.01 |
ENSRNOT00000024922
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr3_+_113918629 | 0.01 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr7_+_64768742 | 0.01 |
ENSRNOT00000005545
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr9_+_92681078 | 0.01 |
ENSRNOT00000034152
|
Sp100
|
SP100 nuclear antigen |
| chr8_-_22086095 | 0.01 |
ENSRNOT00000068598
|
Zglp1
|
zinc finger, GATA-like protein 1 |
| chr10_-_16731898 | 0.01 |
ENSRNOT00000028186
|
Crebrf
|
CREB3 regulatory factor |
| chr3_+_9366053 | 0.01 |
ENSRNOT00000073999
|
Fibcd1l1
|
fibrinogen C domain containing 1-like 1 |
| chr3_+_154910626 | 0.01 |
ENSRNOT00000079515
|
Ralgapb
|
Ral GTPase activating protein non-catalytic beta subunit |
| chr6_-_26680284 | 0.01 |
ENSRNOT00000039709
|
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr2_+_8732932 | 0.01 |
ENSRNOT00000072849
|
Arrdc3
|
arrestin domain containing 3 |
| chr10_+_45322248 | 0.01 |
ENSRNOT00000047268
|
Trim11
|
tripartite motif-containing 11 |
| chr9_+_100489852 | 0.01 |
ENSRNOT00000022854
|
Ppp1r7
|
protein phosphatase 1, regulatory subunit 7 |
| chr9_+_66888393 | 0.01 |
ENSRNOT00000023536
|
Carf
|
calcium responsive transcription factor |
| chr2_-_120316357 | 0.01 |
ENSRNOT00000065469
|
Ccdc39
|
coiled-coil domain containing 39 |
| chr15_-_34693034 | 0.01 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr6_+_73358112 | 0.01 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr18_+_17403407 | 0.01 |
ENSRNOT00000045150
|
RGD1562608
|
similar to KIAA1328 protein |
| chr5_+_128083118 | 0.01 |
ENSRNOT00000079161
|
Zcchc11
|
zinc finger CCHC-type containing 11 |
| chr5_+_64566804 | 0.01 |
ENSRNOT00000073192
|
LOC103690035
|
TGF-beta receptor type-1-like |
| chr10_-_109802739 | 0.01 |
ENSRNOT00000054951
|
Sirt7
|
sirtuin 7 |
| chr1_+_167522895 | 0.01 |
ENSRNOT00000092987
|
Stim1
|
stromal interaction molecule 1 |
| chr12_-_11175917 | 0.01 |
ENSRNOT00000079573
|
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr1_+_266844480 | 0.01 |
ENSRNOT00000027482
|
Taf5
|
TATA-box binding protein associated factor 5 |
| chrX_+_65040775 | 0.01 |
ENSRNOT00000081354
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chrX_+_33884499 | 0.01 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr9_-_11027506 | 0.01 |
ENSRNOT00000071107
|
Chaf1a
|
chromatin assembly factor 1 subunit A |
| chr1_+_64928503 | 0.01 |
ENSRNOT00000086274
|
Vom2r80
|
vomeronasal 2 receptor, 80 |
| chr4_-_173640684 | 0.01 |
ENSRNOT00000010728
|
Rergl
|
RERG like |
| chr1_+_42169501 | 0.01 |
ENSRNOT00000025477
ENSRNOT00000092791 |
Vip
|
vasoactive intestinal peptide |
| chr1_-_126211439 | 0.01 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr15_-_100303807 | 0.01 |
ENSRNOT00000041344
|
LOC102548369
|
40S ribosomal protein S16-like |
| chr14_-_20920286 | 0.01 |
ENSRNOT00000004391
|
Slc4a4
|
solute carrier family 4 member 4 |
| chr7_+_66742987 | 0.01 |
ENSRNOT00000045130
|
Mon2
|
MON2 homolog, regulator of endosome-to-Golgi trafficking |
| chr8_-_52828134 | 0.01 |
ENSRNOT00000007736
|
Rbm7
|
RNA binding motif protein 7 |
| chr15_+_4240203 | 0.01 |
ENSRNOT00000010178
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr6_+_8218696 | 0.01 |
ENSRNOT00000083470
|
Ppm1b
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr7_+_140829076 | 0.01 |
ENSRNOT00000086179
|
Spats2
|
spermatogenesis associated, serine-rich 2 |
| chr5_+_167142182 | 0.00 |
ENSRNOT00000024054
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr13_-_94289333 | 0.00 |
ENSRNOT00000078195
|
Pld5
|
phospholipase D family, member 5 |
| chr3_-_14643897 | 0.00 |
ENSRNOT00000082008
ENSRNOT00000025983 |
Ggta1
|
glycoprotein, alpha-galactosyltransferase 1 |
| chr17_-_43770561 | 0.00 |
ENSRNOT00000088408
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr4_+_158088505 | 0.00 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr9_-_7891514 | 0.00 |
ENSRNOT00000072684
|
Pot1b
|
protection of telomeres 1B |
| chr2_+_188745503 | 0.00 |
ENSRNOT00000056652
|
Shc1
|
SHC adaptor protein 1 |
| chr2_-_35104963 | 0.00 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr4_+_127164453 | 0.00 |
ENSRNOT00000017889
|
Kbtbd8
|
kelch repeat and BTB domain containing 8 |
| chr4_-_6978073 | 0.00 |
ENSRNOT00000045311
|
Nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr1_+_277355619 | 0.00 |
ENSRNOT00000022788
|
Nhlrc2
|
NHL repeat containing 2 |
| chrX_+_104734082 | 0.00 |
ENSRNOT00000005020
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr7_+_58814805 | 0.00 |
ENSRNOT00000005909
|
Tspan8
|
tetraspanin 8 |
| chr15_-_93748742 | 0.00 |
ENSRNOT00000093370
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr4_-_157486844 | 0.00 |
ENSRNOT00000038281
ENSRNOT00000022874 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr2_-_188745144 | 0.00 |
ENSRNOT00000055533
|
Cks1b
|
CDC28 protein kinase regulatory subunit 1B |
| chr10_-_107386072 | 0.00 |
ENSRNOT00000004290
|
Timp2
|
TIMP metallopeptidase inhibitor 2 |
| chr7_+_132378273 | 0.00 |
ENSRNOT00000010990
|
LOC690142
|
hypothetical protein LOC690142 |
| chr2_+_54466280 | 0.00 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr6_+_52459946 | 0.00 |
ENSRNOT00000073277
ENSRNOT00000013474 |
Atxn7l1
|
ataxin 7-like 1 |
| chr18_+_29999290 | 0.00 |
ENSRNOT00000027372
|
Pcdha4
|
protocadherin alpha 4 |
| chr14_-_115391652 | 0.00 |
ENSRNOT00000002094
|
Chac2
|
ChaC cation transport regulator 2 |
| chr6_+_37144787 | 0.00 |
ENSRNOT00000075503
|
Rad51ap2
|
RAD51 associated protein 2 |
| chr2_+_200572502 | 0.00 |
ENSRNOT00000074666
|
Zfp697
|
zinc finger protein 697 |
| chr3_+_154905141 | 0.00 |
ENSRNOT00000088748
ENSRNOT00000020556 |
Ralgapb
|
Ral GTPase activating protein non-catalytic beta subunit |
| chrX_+_106306795 | 0.00 |
ENSRNOT00000073661
|
Gprasp1
|
G protein-coupled receptor associated sorting protein 1 |
| chr2_-_79078258 | 0.00 |
ENSRNOT00000031170
|
Fbxl7
|
F-box and leucine-rich repeat protein 7 |
| chr1_-_198652674 | 0.00 |
ENSRNOT00000023706
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr2_-_140334912 | 0.00 |
ENSRNOT00000015067
|
Elf2
|
E74-like factor 2 |
| chr9_+_52023295 | 0.00 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr12_-_40590361 | 0.00 |
ENSRNOT00000067503
|
Tmem116
|
transmembrane protein 116 |
| chr17_+_2690062 | 0.00 |
ENSRNOT00000024937
|
Olr1653
|
olfactory receptor 1653 |
| chr15_-_39705208 | 0.00 |
ENSRNOT00000015412
|
Phf11
|
PHD finger protein 11 |
| chr16_+_50152008 | 0.00 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr6_-_26314844 | 0.00 |
ENSRNOT00000006658
|
Zfp512
|
zinc finger protein 512 |
| chr4_+_56981283 | 0.00 |
ENSRNOT00000010989
|
Tspan33
|
tetraspanin 33 |
| chr2_+_206064394 | 0.00 |
ENSRNOT00000077739
|
Syt6
|
synaptotagmin 6 |
| chr11_-_4397361 | 0.00 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr8_+_1459526 | 0.00 |
ENSRNOT00000034503
|
Kbtbd3
|
kelch repeat and BTB domain containing 3 |
| chr8_+_106816152 | 0.00 |
ENSRNOT00000047613
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr3_+_44025300 | 0.00 |
ENSRNOT00000006319
|
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr12_+_13715843 | 0.00 |
ENSRNOT00000042459
|
Actb
|
actin, beta |
| chr10_-_84920886 | 0.00 |
ENSRNOT00000068083
|
Sp2
|
Sp2 transcription factor |
| chr8_-_60570058 | 0.00 |
ENSRNOT00000009169
|
Scaper
|
S-phase cyclin A-associated protein in the ER |
| chr10_-_70337532 | 0.00 |
ENSRNOT00000055963
|
Slfn13
|
schlafen family member 13 |
| chr4_+_182483194 | 0.00 |
ENSRNOT00000002528
|
Far2
|
fatty acyl CoA reductase 2 |
| chr2_+_52431601 | 0.00 |
ENSRNOT00000023554
|
Hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chr14_-_66978499 | 0.00 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr3_+_67538289 | 0.00 |
ENSRNOT00000009839
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr14_-_21553949 | 0.00 |
ENSRNOT00000031642
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chr17_+_78876111 | 0.00 |
ENSRNOT00000057895
|
Rpl34-ps1
|
ribosomal protein L34, pseudogene 1 |
| chr9_+_81689802 | 0.00 |
ENSRNOT00000021432
|
Vil1
|
villin 1 |
| chr2_-_250805445 | 0.00 |
ENSRNOT00000055362
|
Clca4l
|
chloride channel calcium activated 4-like |
| chr8_+_130581062 | 0.00 |
ENSRNOT00000014323
|
Fam198a
|
family with sequence similarity 198, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd11_Cdx1_Hoxc11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.0 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.0 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.0 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) regulation of cellular response to drug(GO:2001038) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |