Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
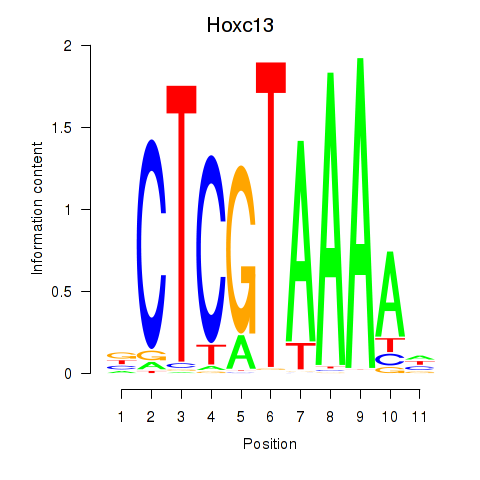
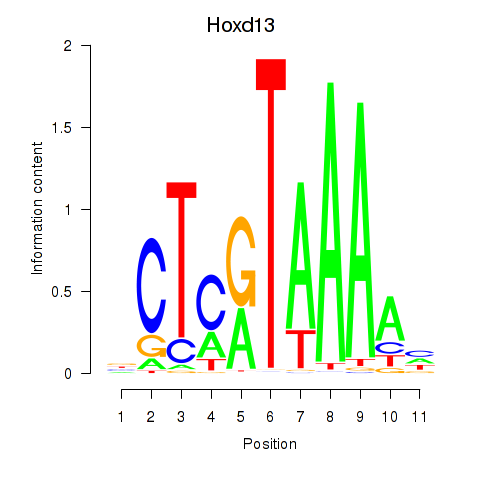
Results for Hoxc13_Hoxd13
Z-value: 0.35
Transcription factors associated with Hoxc13_Hoxd13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc13
|
ENSRNOG00000028679 | homeobox C13 |
|
Hoxd13
|
ENSRNOG00000001588 | homeo box D13 |
Activity profile of Hoxc13_Hoxd13 motif
Sorted Z-values of Hoxc13_Hoxd13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_155955173 | 0.21 |
ENSRNOT00000079345
|
AABR07004776.1
|
|
| chr9_+_67699379 | 0.15 |
ENSRNOT00000091237
ENSRNOT00000088183 |
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr3_-_165700489 | 0.14 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr11_-_29710849 | 0.13 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr4_-_159192526 | 0.11 |
ENSRNOT00000026731
|
Kcna1
|
potassium voltage-gated channel subfamily A member 1 |
| chr10_-_87954055 | 0.11 |
ENSRNOT00000018048
|
Krt34
|
keratin 34 |
| chr5_-_68139199 | 0.09 |
ENSRNOT00000009987
|
Plppr1
|
phospholipid phosphatase related 1 |
| chrX_-_106607352 | 0.07 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chr10_-_88019967 | 0.06 |
ENSRNOT00000047580
|
Krt36
|
keratin 36 |
| chr2_-_220838905 | 0.06 |
ENSRNOT00000078914
|
AABR07013065.1
|
|
| chr6_-_41039437 | 0.06 |
ENSRNOT00000005774
|
Trib2
|
tribbles pseudokinase 2 |
| chr4_-_82271893 | 0.06 |
ENSRNOT00000075005
|
Hoxa7
|
homeobox A7 |
| chr7_-_143228060 | 0.06 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr18_-_40218225 | 0.05 |
ENSRNOT00000004723
|
Pggt1b
|
protein geranylgeranyltransferase type 1 subunit beta |
| chr8_+_96551245 | 0.05 |
ENSRNOT00000039850
|
Bcl2a1
|
BCL2-related protein A1 |
| chr5_-_154332940 | 0.05 |
ENSRNOT00000014458
|
Pithd1
|
PITH domain containing 1 |
| chr3_+_46185360 | 0.04 |
ENSRNOT00000051344
|
LOC100361645
|
LRRGT00075-like |
| chr10_+_108132105 | 0.04 |
ENSRNOT00000072534
|
Cbx2
|
chromobox 2 |
| chr10_-_87248572 | 0.04 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr1_+_236031988 | 0.04 |
ENSRNOT00000016164
|
Pcsk5
|
proprotein convertase subtilisin/kexin type 5 |
| chr20_-_27578244 | 0.04 |
ENSRNOT00000000708
|
Fam26d
|
family with sequence similarity 26, member D |
| chr16_+_2706428 | 0.04 |
ENSRNOT00000077117
|
Il17rd
|
interleukin 17 receptor D |
| chr7_-_143453544 | 0.03 |
ENSRNOT00000034450
ENSRNOT00000083956 |
Krt1
Krt5
|
keratin 1 keratin 5 |
| chr10_+_94192556 | 0.03 |
ENSRNOT00000074007
ENSRNOT00000093039 |
Ace3
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 3 |
| chr7_-_3123297 | 0.03 |
ENSRNOT00000061878
|
Rab5b
|
RAB5B, member RAS oncogene family |
| chr19_+_37025375 | 0.03 |
ENSRNOT00000048385
|
Ces2g
|
carboxylesterase 2G |
| chr18_-_28017925 | 0.03 |
ENSRNOT00000075420
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr4_-_82186190 | 0.03 |
ENSRNOT00000071729
|
LOC100911622
|
homeobox protein Hox-A7-like |
| chr17_-_84247038 | 0.03 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr15_-_11912806 | 0.03 |
ENSRNOT00000068171
ENSRNOT00000008759 ENSRNOT00000049771 |
Slc4a7
|
solute carrier family 4 member 7 |
| chr9_+_66058047 | 0.03 |
ENSRNOT00000071819
|
Cdk15
|
cyclin-dependent kinase 15 |
| chr1_+_154131926 | 0.03 |
ENSRNOT00000035257
|
LOC102549852
|
ferritin light chain 1-like |
| chr8_-_39734594 | 0.02 |
ENSRNOT00000076872
|
Slc37a2
|
solute carrier family 37 member 2 |
| chr9_+_65534704 | 0.02 |
ENSRNOT00000016730
|
Cflar
|
CASP8 and FADD-like apoptosis regulator |
| chr13_-_80702315 | 0.02 |
ENSRNOT00000004577
|
Fmo4
|
flavin containing monooxygenase 4 |
| chr11_+_34316295 | 0.02 |
ENSRNOT00000081834
|
Sim2
|
single-minded family bHLH transcription factor 2 |
| chr6_+_37144787 | 0.02 |
ENSRNOT00000075503
|
Rad51ap2
|
RAD51 associated protein 2 |
| chr11_-_782954 | 0.02 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr2_-_43067956 | 0.02 |
ENSRNOT00000090616
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr13_-_45040593 | 0.02 |
ENSRNOT00000004908
|
Lct
|
lactase |
| chr6_+_69971227 | 0.02 |
ENSRNOT00000075349
|
Foxg1
|
forkhead box G1 |
| chr15_+_41643541 | 0.02 |
ENSRNOT00000019646
|
Arl11
|
ADP-ribosylation factor like GTPase 11 |
| chr20_+_37876650 | 0.02 |
ENSRNOT00000001054
|
Gja1
|
gap junction protein, alpha 1 |
| chr14_+_17210733 | 0.02 |
ENSRNOT00000003075
|
Cxcl10
|
C-X-C motif chemokine ligand 10 |
| chr6_+_123034304 | 0.02 |
ENSRNOT00000078938
|
Cpg1
|
candidate plasticity gene 1 |
| chr10_-_88000423 | 0.02 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr12_-_21760292 | 0.02 |
ENSRNOT00000059592
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr5_-_146795866 | 0.02 |
ENSRNOT00000065640
|
Tlr12
|
toll-like receptor 12 |
| chr2_-_9472384 | 0.02 |
ENSRNOT00000084295
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr5_-_16140896 | 0.01 |
ENSRNOT00000029503
|
Xkr4
|
XK related 4 |
| chr5_+_157327657 | 0.01 |
ENSRNOT00000022847
|
Pla2g2e
|
phospholipase A2, group IIE |
| chr17_+_70262363 | 0.01 |
ENSRNOT00000048933
|
Fam208b
|
family with sequence similarity 208, member B |
| chr10_-_61232331 | 0.01 |
ENSRNOT00000065045
|
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr3_+_11476883 | 0.01 |
ENSRNOT00000072241
|
Naif1
|
nuclear apoptosis inducing factor 1 |
| chr10_-_87437455 | 0.01 |
ENSRNOT00000051080
|
Krt39
|
keratin 39 |
| chr8_+_122197027 | 0.01 |
ENSRNOT00000013050
|
Ubp1
|
upstream binding protein 1 (LBP-1a) |
| chr2_-_173800761 | 0.01 |
ENSRNOT00000039524
|
Wdr49
|
WD repeat domain 49 |
| chr3_-_120373500 | 0.01 |
ENSRNOT00000067727
|
Nphp1
|
nephrocystin 1 |
| chr9_+_79659251 | 0.01 |
ENSRNOT00000021656
|
Xrcc5
|
X-ray repair cross complementing 5 |
| chr11_-_61470046 | 0.01 |
ENSRNOT00000073436
|
LOC102553099
|
N-alpha-acetyltransferase 50-like |
| chr2_+_54466280 | 0.01 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr19_-_19896421 | 0.01 |
ENSRNOT00000020793
|
Heatr3
|
HEAT repeat containing 3 |
| chr20_+_11655967 | 0.01 |
ENSRNOT00000064565
|
LOC690478
|
similar to keratin associated protein 10-7 |
| chr9_-_52238564 | 0.01 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr12_+_6403940 | 0.01 |
ENSRNOT00000083484
|
B3glct
|
beta 3-glucosyltransferase |
| chrX_+_20216587 | 0.01 |
ENSRNOT00000073114
|
AABR07037412.2
|
FYVE, RhoGEF and PH domain-containing protein 1 |
| chrX_-_77675487 | 0.01 |
ENSRNOT00000042799
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr7_+_107406962 | 0.01 |
ENSRNOT00000093254
|
Phf20l1
|
PHD finger protein 20-like protein 1 |
| chr4_-_2201749 | 0.01 |
ENSRNOT00000089327
|
Lmbr1
|
limb development membrane protein 1 |
| chr7_-_140437467 | 0.01 |
ENSRNOT00000087181
|
Fkbp11
|
FK506 binding protein 11 |
| chr2_+_210880777 | 0.01 |
ENSRNOT00000026237
|
Gnat2
|
G protein subunit alpha transducin 2 |
| chr11_+_45401403 | 0.01 |
ENSRNOT00000002240
|
Tbc1d23
|
TBC1 domain family, member 23 |
| chr16_-_37177033 | 0.01 |
ENSRNOT00000014015
|
Fbxo8
|
F-box protein 8 |
| chr10_+_84135116 | 0.01 |
ENSRNOT00000031035
|
Hoxb7
|
homeo box B7 |
| chr8_+_115546712 | 0.01 |
ENSRNOT00000079801
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr1_-_43638161 | 0.01 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr19_-_19315357 | 0.00 |
ENSRNOT00000018888
|
Cyld
|
CYLD lysine 63 deubiquitinase |
| chr1_-_219745654 | 0.00 |
ENSRNOT00000054848
|
LOC689065
|
hypothetical protein LOC689065 |
| chr9_-_43454078 | 0.00 |
ENSRNOT00000023550
|
Tmem131
|
transmembrane protein 131 |
| chr6_-_105261549 | 0.00 |
ENSRNOT00000009122
|
Synj2bp
|
synaptojanin 2 binding protein |
| chr17_+_87274944 | 0.00 |
ENSRNOT00000073718
|
Etl4
|
enhancer trap locus 4 |
| chr2_+_180936688 | 0.00 |
ENSRNOT00000016039
|
Asic5
|
acid sensing ion channel subunit family member 5 |
| chr8_+_115546893 | 0.00 |
ENSRNOT00000018932
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr1_+_261753364 | 0.00 |
ENSRNOT00000076659
|
R3hcc1l
|
R3H domain and coiled-coil containing 1-like |
| chr4_-_78879294 | 0.00 |
ENSRNOT00000084543
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr3_+_110574417 | 0.00 |
ENSRNOT00000031231
|
Disp2
|
dispatched RND transporter family member 2 |
| chr8_+_17421557 | 0.00 |
ENSRNOT00000030569
|
Chordc1
|
cysteine and histidine rich domain containing 1 |
| chr14_-_21553949 | 0.00 |
ENSRNOT00000031642
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chr5_-_125345726 | 0.00 |
ENSRNOT00000011244
|
LOC688684
|
similar to 60S ribosomal protein L32 |
| chr1_-_225077079 | 0.00 |
ENSRNOT00000074747
|
Uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr10_+_38918748 | 0.00 |
ENSRNOT00000009999
|
Kif3a
|
kinesin family member 3a |
| chr13_+_27238767 | 0.00 |
ENSRNOT00000003495
|
Serpinb11
|
serpin family B member 11 |
| chr3_+_160047296 | 0.00 |
ENSRNOT00000051590
|
Pkig
|
cAMP-dependent protein kinase inhibitor gamma |
| chr6_+_99625306 | 0.00 |
ENSRNOT00000008573
|
Plekhg3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr7_-_18634079 | 0.00 |
ENSRNOT00000010031
|
Angptl4
|
angiopoietin-like 4 |
| chr10_-_56368360 | 0.00 |
ENSRNOT00000019674
|
Slc35g3
|
solute carrier family 35, member G3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc13_Hoxd13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |