Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Hoxb8_Pdx1
Z-value: 0.31
Transcription factors associated with Hoxb8_Pdx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
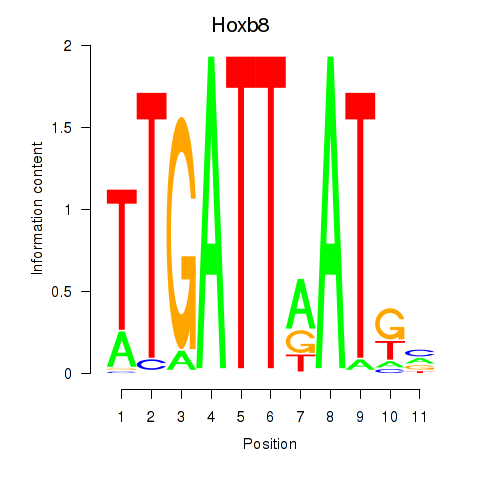
Hoxb8
|
ENSRNOG00000007585 | homeo box B8 |
|
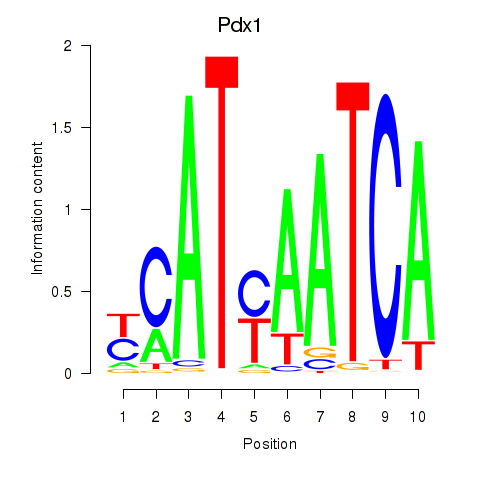
Pdx1
|
ENSRNOG00000046458 | pancreatic and duodenal homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb8 | rn6_v1_chr10_+_84131495_84131495 | 0.13 | 8.4e-01 | Click! |
Activity profile of Hoxb8_Pdx1 motif
Sorted Z-values of Hoxb8_Pdx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_82133605 | 0.20 |
ENSRNOT00000008023
|
NEWGENE_2813
|
homeobox A2 |
| chr10_+_84200880 | 0.13 |
ENSRNOT00000011213
|
Hoxb2
|
homeobox B2 |
| chr7_+_37812831 | 0.09 |
ENSRNOT00000005910
|
Btg1
|
BTG anti-proliferation factor 1 |
| chrX_+_136460215 | 0.09 |
ENSRNOT00000093538
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr14_+_39368530 | 0.08 |
ENSRNOT00000084367
|
Cox7b2
|
cytochrome c oxidase subunit 7B2 |
| chr3_+_159368273 | 0.08 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr5_-_2803855 | 0.08 |
ENSRNOT00000009490
|
LOC297756
|
ribosomal protein S8-like |
| chr11_+_80358211 | 0.08 |
ENSRNOT00000002519
|
Sst
|
somatostatin |
| chr7_-_145062956 | 0.07 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr3_+_160908769 | 0.07 |
ENSRNOT00000030054
|
Sys1
|
Sys1 golgi trafficking protein |
| chr10_-_87407634 | 0.07 |
ENSRNOT00000016657
|
Krt23
|
keratin 23 |
| chr11_+_53140599 | 0.07 |
ENSRNOT00000083438
|
Bbx
|
BBX, HMG-box containing |
| chr16_-_11214948 | 0.07 |
ENSRNOT00000023369
|
LOC690468
|
similar to 60S ribosomal protein L38 |
| chrX_+_124894466 | 0.07 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr14_+_32257805 | 0.07 |
ENSRNOT00000039626
|
AABR07014804.1
|
|
| chr11_+_17538063 | 0.06 |
ENSRNOT00000031889
ENSRNOT00000090878 |
Chodl
|
chondrolectin |
| chr1_-_141655417 | 0.06 |
ENSRNOT00000068084
|
Ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr14_-_62595854 | 0.05 |
ENSRNOT00000003024
|
Yipf7
|
Yip1 domain family, member 7 |
| chr1_-_155955173 | 0.05 |
ENSRNOT00000079345
|
AABR07004776.1
|
|
| chr14_-_14390699 | 0.05 |
ENSRNOT00000046639
|
Anxa3
|
annexin A3 |
| chr1_+_264893162 | 0.05 |
ENSRNOT00000021714
|
Tlx1
|
T-cell leukemia, homeobox 1 |
| chrX_-_123486814 | 0.05 |
ENSRNOT00000016993
|
RGD1564541
|
similar to hypothetical protein FLJ22965 |
| chr19_-_37427989 | 0.05 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr9_+_53440272 | 0.05 |
ENSRNOT00000059159
|
LOC685203
|
hypothetical protein LOC685203 |
| chr16_-_7412150 | 0.05 |
ENSRNOT00000047252
|
RGD1564325
|
similar to ribosomal protein S24 |
| chr2_+_212247451 | 0.05 |
ENSRNOT00000027813
|
Vav3
|
vav guanine nucleotide exchange factor 3 |
| chr5_-_22769907 | 0.05 |
ENSRNOT00000047805
ENSRNOT00000076167 ENSRNOT00000076507 ENSRNOT00000076113 ENSRNOT00000083779 |
Asph
|
aspartate-beta-hydroxylase |
| chr3_+_114900343 | 0.05 |
ENSRNOT00000068129
|
Sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr3_-_161131812 | 0.05 |
ENSRNOT00000067008
|
Wfdc11
|
WAP four-disulfide core domain 11 |
| chrX_+_119390013 | 0.05 |
ENSRNOT00000074269
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr14_-_72970329 | 0.05 |
ENSRNOT00000006325
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr8_-_104155775 | 0.05 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chrX_+_35869538 | 0.04 |
ENSRNOT00000058947
|
Ppef1
|
protein phosphatase with EF-hand domain 1 |
| chrX_+_112311251 | 0.04 |
ENSRNOT00000086698
|
AABR07040855.1
|
|
| chr5_-_12172009 | 0.04 |
ENSRNOT00000061903
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chrX_+_124894706 | 0.04 |
ENSRNOT00000066677
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr4_-_159192526 | 0.04 |
ENSRNOT00000026731
|
Kcna1
|
potassium voltage-gated channel subfamily A member 1 |
| chr10_-_18506337 | 0.04 |
ENSRNOT00000043036
|
Gabrp
|
gamma-aminobutyric acid type A receptor pi subunit |
| chr7_-_1738642 | 0.04 |
ENSRNOT00000075554
|
AABR07055356.1
|
|
| chr1_-_67094567 | 0.04 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr7_+_130542202 | 0.04 |
ENSRNOT00000079501
ENSRNOT00000045647 |
Acr
|
acrosin |
| chr5_-_164348713 | 0.04 |
ENSRNOT00000087044
|
LOC500584
|
similar to casein kinase 1, gamma 3 isoform 2 |
| chr16_-_52127591 | 0.04 |
ENSRNOT00000033652
|
Triml2
|
tripartite motif family-like 2 |
| chr1_+_15834779 | 0.04 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr9_-_4685383 | 0.04 |
ENSRNOT00000061896
|
Sult1c2
|
sulfotransferase family 1C member 2 |
| chr4_+_166601960 | 0.04 |
ENSRNOT00000073394
|
AABR07062266.1
|
|
| chr8_+_99632803 | 0.04 |
ENSRNOT00000087190
|
Plscr1
|
phospholipid scramblase 1 |
| chr3_-_45169118 | 0.04 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr14_-_91979500 | 0.04 |
ENSRNOT00000073892
|
Ddc
|
dopa decarboxylase |
| chr8_+_130350510 | 0.04 |
ENSRNOT00000073376
|
Ss18l2
|
SS18 like 2 |
| chr8_-_49227273 | 0.04 |
ENSRNOT00000050878
|
Atp5l
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G |
| chr8_-_92942076 | 0.03 |
ENSRNOT00000056937
|
Fam46a
|
family with sequence similarity 46, member A |
| chr8_+_117906014 | 0.03 |
ENSRNOT00000056180
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chrX_-_45284341 | 0.03 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chr4_-_132111079 | 0.03 |
ENSRNOT00000013719
|
Eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr15_-_44442875 | 0.03 |
ENSRNOT00000017939
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr9_+_15313523 | 0.03 |
ENSRNOT00000070805
|
Tomm6
|
translocase of outer mitochondrial membrane 6 |
| chr18_-_85980833 | 0.03 |
ENSRNOT00000058179
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr8_-_77599781 | 0.03 |
ENSRNOT00000087980
|
Aqp9
|
aquaporin 9 |
| chr15_+_1054937 | 0.03 |
ENSRNOT00000008154
|
AABR07016841.1
|
|
| chr14_+_7949239 | 0.03 |
ENSRNOT00000044617
|
Ndufb4l1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4-like 1 |
| chr15_+_28439317 | 0.03 |
ENSRNOT00000060519
|
LOC690384
|
similar to ribosomal protein L31 |
| chrX_+_105134498 | 0.03 |
ENSRNOT00000002058
|
Tmem35
|
transmembrane protein 35 |
| chr5_-_58106706 | 0.03 |
ENSRNOT00000019561
|
Dctn3
|
dynactin subunit 3 |
| chrX_+_31984612 | 0.03 |
ENSRNOT00000005181
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr17_+_69960160 | 0.03 |
ENSRNOT00000023887
|
Ucn3
|
urocortin 3 |
| chrX_-_142248369 | 0.03 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr4_-_181348799 | 0.03 |
ENSRNOT00000033743
|
LOC690784
|
hypothetical protein LOC690783 |
| chr10_-_87521514 | 0.03 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr2_+_210045161 | 0.03 |
ENSRNOT00000024455
|
Slc16a4
|
solute carrier family 16, member 4 |
| chr4_-_82263302 | 0.03 |
ENSRNOT00000082933
|
LOC100909604
|
homeobox protein Hox-A6-like |
| chrX_+_54553878 | 0.03 |
ENSRNOT00000040231
|
Samt3
|
spermatogenesis associated multipass transmembrane protein 3 |
| chr8_-_77398156 | 0.03 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr10_-_87954055 | 0.03 |
ENSRNOT00000018048
|
Krt34
|
keratin 34 |
| chr13_-_36022197 | 0.03 |
ENSRNOT00000091280
|
Cfap221
|
cilia and flagella associated protein 221 |
| chr6_-_127620296 | 0.03 |
ENSRNOT00000012577
|
Serpina1
|
serpin family A member 1 |
| chr19_+_25983169 | 0.03 |
ENSRNOT00000004404
|
Syce2
|
synaptonemal complex central element protein 2 |
| chr4_-_58250798 | 0.03 |
ENSRNOT00000048436
|
Klf14
|
Kruppel-like factor 14 |
| chr14_-_72122158 | 0.03 |
ENSRNOT00000064495
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr6_-_51018050 | 0.03 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr2_-_259382765 | 0.03 |
ENSRNOT00000091407
|
St6galnac3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr2_-_123972356 | 0.03 |
ENSRNOT00000023348
|
Il21
|
interleukin 21 |
| chr11_+_88376256 | 0.03 |
ENSRNOT00000081776
|
Vpreb1
|
pre-B lymphocyte 1 |
| chr3_-_47025128 | 0.03 |
ENSRNOT00000011682
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr3_-_61581598 | 0.03 |
ENSRNOT00000002156
|
Evx2
|
even-skipped homeobox 2 |
| chr1_-_1889236 | 0.03 |
ENSRNOT00000087944
|
AABR07000159.2
|
|
| chr2_-_252451999 | 0.03 |
ENSRNOT00000021861
|
Dnase2b
|
deoxyribonuclease 2 beta |
| chr8_-_63291966 | 0.03 |
ENSRNOT00000077762
|
AABR07070278.1
|
|
| chr7_+_125288081 | 0.03 |
ENSRNOT00000085216
|
Parvg
|
parvin, gamma |
| chr3_+_31802999 | 0.03 |
ENSRNOT00000041305
|
AABR07051996.1
|
|
| chr19_+_43163129 | 0.03 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr3_-_23020441 | 0.02 |
ENSRNOT00000017651
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr16_-_31301880 | 0.02 |
ENSRNOT00000084847
ENSRNOT00000083943 |
AABR07025272.1
|
|
| chr3_-_151363115 | 0.02 |
ENSRNOT00000073492
|
Eif6
|
eukaryotic translation initiation factor 6 |
| chr3_+_118317761 | 0.02 |
ENSRNOT00000012700
|
Fgf7
|
fibroblast growth factor 7 |
| chr16_-_29936307 | 0.02 |
ENSRNOT00000088707
|
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr5_-_128446725 | 0.02 |
ENSRNOT00000011272
|
Btf3l4
|
basic transcription factor 3-like 4 |
| chr1_+_85485289 | 0.02 |
ENSRNOT00000026182
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr5_+_133864798 | 0.02 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr7_-_144837583 | 0.02 |
ENSRNOT00000055289
|
Cbx5
|
chromobox 5 |
| chrX_-_83864150 | 0.02 |
ENSRNOT00000073734
|
Obp1f
|
odorant binding protein I f |
| chr14_+_22107416 | 0.02 |
ENSRNOT00000060179
ENSRNOT00000080114 |
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr1_+_46978458 | 0.02 |
ENSRNOT00000024211
|
Gtf2h5
|
general transcription factor IIH subunit 5 |
| chr13_+_108642156 | 0.02 |
ENSRNOT00000004783
|
Smyd2
|
SET and MYND domain containing 2 |
| chr14_+_79013807 | 0.02 |
ENSRNOT00000091068
|
Bloc1s4
|
biogenesis of lysosomal organelles complex 1 subunit 4 |
| chr18_-_81682206 | 0.02 |
ENSRNOT00000058219
|
LOC100359752
|
hypothetical protein LOC100359752 |
| chr4_-_162856663 | 0.02 |
ENSRNOT00000077791
|
Clec2e
|
C-type lectin domain family 2, member E |
| chr3_+_51883559 | 0.02 |
ENSRNOT00000007197
|
Csrnp3
|
cysteine and serine rich nuclear protein 3 |
| chr19_+_34616768 | 0.02 |
ENSRNOT00000051194
|
Rps27a-ps6
|
ribosomal protein S27a, pseudogene 6 |
| chr7_+_117519075 | 0.02 |
ENSRNOT00000029768
|
Scx
|
scleraxis bHLH transcription factor |
| chr16_+_61926586 | 0.02 |
ENSRNOT00000058884
|
AABR07026059.1
|
|
| chr6_-_75670135 | 0.02 |
ENSRNOT00000007300
|
Snx6
|
sorting nexin 6 |
| chr10_+_46018659 | 0.02 |
ENSRNOT00000040865
ENSRNOT00000004357 |
Mprip
|
myosin phosphatase Rho interacting protein |
| chr4_+_117594805 | 0.02 |
ENSRNOT00000074924
|
LOC103690067
|
EKC/KEOPS complex subunit Tprkb |
| chr4_-_170860225 | 0.02 |
ENSRNOT00000007577
|
Mgp
|
matrix Gla protein |
| chr17_-_71897972 | 0.02 |
ENSRNOT00000065942
|
Sfmbt2
|
Scm-like with four mbt domains 2 |
| chr3_-_134696654 | 0.02 |
ENSRNOT00000006454
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr3_-_176548208 | 0.02 |
ENSRNOT00000084339
|
Chrna4
|
cholinergic receptor nicotinic alpha 4 subunit |
| chr20_-_25826658 | 0.02 |
ENSRNOT00000057950
ENSRNOT00000084660 |
Ctnna3
|
catenin alpha 3 |
| chr1_+_84280945 | 0.02 |
ENSRNOT00000057190
|
Sertad3
|
SERTA domain containing 3 |
| chr4_-_84131030 | 0.02 |
ENSRNOT00000012196
|
Cpvl
|
carboxypeptidase, vitellogenic-like |
| chr14_+_42007312 | 0.02 |
ENSRNOT00000063985
|
Atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr2_+_240461505 | 0.02 |
ENSRNOT00000019507
|
Bdh2
|
3-hydroxybutyrate dehydrogenase 2 |
| chr18_-_36893797 | 0.02 |
ENSRNOT00000043544
|
Gpr151
|
G protein-coupled receptor 151 |
| chr12_-_13133219 | 0.02 |
ENSRNOT00000092292
|
Daglb
|
diacylglycerol lipase, beta |
| chr9_+_67699379 | 0.02 |
ENSRNOT00000091237
ENSRNOT00000088183 |
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr4_+_57925323 | 0.02 |
ENSRNOT00000085798
|
Cpa5
|
carboxypeptidase A5 |
| chr2_+_147496229 | 0.02 |
ENSRNOT00000022105
|
Tm4sf4
|
transmembrane 4 L six family member 4 |
| chr1_-_101095594 | 0.02 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr2_-_203680083 | 0.02 |
ENSRNOT00000021268
|
Cd2
|
Cd2 molecule |
| chr17_-_90218013 | 0.02 |
ENSRNOT00000072803
|
Impad1
|
inositol monophosphatase domain containing 1 |
| chr16_+_61130755 | 0.02 |
ENSRNOT00000042609
|
AABR07026048.1
|
|
| chr8_-_27973980 | 0.02 |
ENSRNOT00000037757
|
Bin2a
|
beta-galactosidase-like protein |
| chr17_-_15519060 | 0.02 |
ENSRNOT00000093624
|
Aspnl1
|
asporin-like 1 |
| chr9_-_121972055 | 0.02 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr2_-_210738378 | 0.02 |
ENSRNOT00000025746
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr1_+_61374379 | 0.02 |
ENSRNOT00000015343
|
Zfp53
|
zinc finger protein 53 |
| chr9_+_94928489 | 0.02 |
ENSRNOT00000087366
ENSRNOT00000085770 ENSRNOT00000024733 |
Sag
|
S-antigen visual arrestin |
| chr14_+_19319299 | 0.02 |
ENSRNOT00000086542
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr5_-_100970043 | 0.02 |
ENSRNOT00000013950
|
Zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr5_+_133865331 | 0.02 |
ENSRNOT00000035409
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr20_+_47395014 | 0.02 |
ENSRNOT00000057116
|
Ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr4_-_170740274 | 0.02 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
| chr3_+_4867233 | 0.02 |
ENSRNOT00000006754
|
Rpl7a
|
ribosomal protein L7a |
| chr5_-_50138475 | 0.02 |
ENSRNOT00000011969
|
Slc35a1
|
solute carrier family 35 member A1 |
| chr8_-_128521109 | 0.02 |
ENSRNOT00000034025
|
Scn11a
|
sodium voltage-gated channel alpha subunit 11 |
| chr1_+_98425903 | 0.02 |
ENSRNOT00000024170
|
LOC690483
|
hypothetical protein LOC690483 |
| chr4_+_14039977 | 0.02 |
ENSRNOT00000091249
ENSRNOT00000075878 |
Cd36
|
CD36 molecule |
| chr11_+_86421106 | 0.02 |
ENSRNOT00000002599
|
LOC100362453
|
serine/threonine-protein phosphatase 2A catalytic subunit alpha-like |
| chr4_-_164900857 | 0.02 |
ENSRNOT00000078690
|
Ly49i2
|
Ly49 inhibitory receptor 2 |
| chr5_+_8876044 | 0.02 |
ENSRNOT00000008877
|
Cops5
|
COP9 signalosome subunit 5 |
| chrM_+_11736 | 0.02 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr1_-_276012351 | 0.02 |
ENSRNOT00000045642
|
AABR07007000.1
|
|
| chrX_-_42329232 | 0.02 |
ENSRNOT00000045705
|
LOC100361934
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 |
| chr10_-_88107232 | 0.02 |
ENSRNOT00000019330
ENSRNOT00000076770 |
Krt9
|
keratin 9 |
| chr7_+_121930615 | 0.02 |
ENSRNOT00000091270
ENSRNOT00000033975 |
Tnrc6b
|
trinucleotide repeat containing 6B |
| chr4_-_148761547 | 0.02 |
ENSRNOT00000018050
|
Zfp422
|
zinc finger protein 422 |
| chr5_+_165724027 | 0.02 |
ENSRNOT00000018000
|
Casz1
|
castor zinc finger 1 |
| chr4_-_132740938 | 0.02 |
ENSRNOT00000007185
|
Rybp
|
RING1 and YY1 binding protein |
| chr16_+_70778913 | 0.02 |
ENSRNOT00000019281
|
Hook3
|
hook microtubule-tethering protein 3 |
| chr3_+_61658245 | 0.02 |
ENSRNOT00000033511
|
Hoxd3
|
homeo box D3 |
| chr15_+_18322327 | 0.02 |
ENSRNOT00000009650
|
Fam3d
|
family with sequence similarity 3, member D |
| chr7_-_18180705 | 0.02 |
ENSRNOT00000065954
|
Zfp494
|
zinc finger protein 494 |
| chr20_-_5166252 | 0.02 |
ENSRNOT00000001138
|
Aif1
|
allograft inflammatory factor 1 |
| chr4_-_162230859 | 0.01 |
ENSRNOT00000042872
|
LOC689757
|
similar to osteoclast inhibitory lectin |
| chr3_+_160410757 | 0.01 |
ENSRNOT00000055172
|
Pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr1_+_167870452 | 0.01 |
ENSRNOT00000025027
|
Olr63
|
olfactory receptor 63 |
| chr2_+_62198562 | 0.01 |
ENSRNOT00000087636
|
Zfr
|
zinc finger RNA binding protein |
| chr2_+_166856784 | 0.01 |
ENSRNOT00000035765
|
Otol1
|
otolin 1 |
| chr18_+_27632786 | 0.01 |
ENSRNOT00000073564
ENSRNOT00000078969 |
Reep2
|
receptor accessory protein 2 |
| chr12_-_19592000 | 0.01 |
ENSRNOT00000001855
|
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr2_+_55835151 | 0.01 |
ENSRNOT00000018634
|
Fyb
|
FYN binding protein |
| chr4_-_23119005 | 0.01 |
ENSRNOT00000048061
|
Steap4
|
STEAP4 metalloreductase |
| chr17_-_43537293 | 0.01 |
ENSRNOT00000091749
|
Slc17a3
|
solute carrier family 17 member 3 |
| chr6_-_67084234 | 0.01 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr6_-_10592454 | 0.01 |
ENSRNOT00000020600
|
Pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr4_-_181348038 | 0.01 |
ENSRNOT00000082879
|
LOC690784
|
hypothetical protein LOC690783 |
| chr8_+_132828091 | 0.01 |
ENSRNOT00000008269
|
Ccr9
|
C-C motif chemokine receptor 9 |
| chr2_+_237679792 | 0.01 |
ENSRNOT00000064628
|
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr5_-_25750826 | 0.01 |
ENSRNOT00000022249
|
Fam92a
|
family with sequence similarity 92 member A |
| chr12_-_50213400 | 0.01 |
ENSRNOT00000048541
|
LOC102552378
|
trichohyalin-like |
| chr14_+_45062662 | 0.01 |
ENSRNOT00000059124
|
Tlr10
|
toll-like receptor 10 |
| chrY_-_1398030 | 0.01 |
ENSRNOT00000088719
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr5_+_137243094 | 0.01 |
ENSRNOT00000067237
|
Med8
|
mediator complex subunit 8 |
| chr9_+_67774150 | 0.01 |
ENSRNOT00000091060
|
Icos
|
inducible T-cell co-stimulator |
| chr10_+_91217079 | 0.01 |
ENSRNOT00000004218
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr19_-_43848937 | 0.01 |
ENSRNOT00000044844
|
Ldhd
|
lactate dehydrogenase D |
| chr1_+_100501676 | 0.01 |
ENSRNOT00000043724
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr11_+_66713888 | 0.01 |
ENSRNOT00000003340
|
Fbxo40
|
F-box protein 40 |
| chr15_-_18096039 | 0.01 |
ENSRNOT00000042545
|
Ptgdr
|
prostaglandin D2 receptor |
| chr7_-_29171783 | 0.01 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr16_+_72010106 | 0.01 |
ENSRNOT00000058330
|
Adam5
|
ADAM metallopeptidase domain 5 |
| chr8_-_39734594 | 0.01 |
ENSRNOT00000076872
|
Slc37a2
|
solute carrier family 37 member 2 |
| chr10_-_8498422 | 0.01 |
ENSRNOT00000082332
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr1_-_85517360 | 0.01 |
ENSRNOT00000026114
|
Eid2
|
EP300 interacting inhibitor of differentiation 2 |
| chr3_+_122803772 | 0.01 |
ENSRNOT00000009564
|
Nop56
|
NOP56 ribonucleoprotein |
| chr4_-_50312608 | 0.01 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr2_+_205488182 | 0.01 |
ENSRNOT00000024270
|
Sike1
|
suppressor of IKBKE 1 |
| chr2_-_172459165 | 0.01 |
ENSRNOT00000057473
|
Schip1
|
schwannomin interacting protein 1 |
| chr1_+_248723397 | 0.01 |
ENSRNOT00000072188
|
LOC100911854
|
mannose-binding protein C-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb8_Pdx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.0 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) negative regulation of icosanoid secretion(GO:0032304) regulation of metanephros size(GO:0035566) |
| 0.0 | 0.0 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0004058 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.0 | 0.0 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.0 | GO:0015265 | urea channel activity(GO:0015265) |