Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Hoxb5
Z-value: 0.45
Transcription factors associated with Hoxb5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb5
|
ENSRNOG00000008010 | homeo box B5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb5 | rn6_v1_chr10_+_84152152_84152152 | 0.39 | 5.1e-01 | Click! |
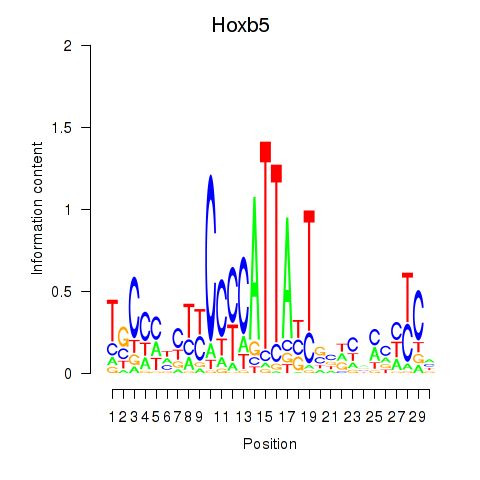
Activity profile of Hoxb5 motif
Sorted Z-values of Hoxb5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_6679878 | 0.58 |
ENSRNOT00000075989
ENSRNOT00000067875 |
Spp1
|
secreted phosphoprotein 1 |
| chr2_+_164549455 | 0.22 |
ENSRNOT00000017151
|
Mlf1
|
myeloid leukemia factor 1 |
| chr20_+_5067330 | 0.15 |
ENSRNOT00000037482
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr5_-_38923095 | 0.13 |
ENSRNOT00000009146
|
AABR07047593.1
|
|
| chr12_-_38782010 | 0.12 |
ENSRNOT00000001813
|
Wdr66
|
WD repeat domain 66 |
| chr4_+_7173961 | 0.11 |
ENSRNOT00000017854
|
Gbx1
|
gastrulation brain homeobox 1 |
| chr10_-_13115294 | 0.11 |
ENSRNOT00000005899
|
NEWGENE_6497122
|
FLYWCH family member 2 |
| chr12_+_2170630 | 0.10 |
ENSRNOT00000071928
|
Pet100
|
PET100 homolog |
| chr10_-_83898527 | 0.10 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr2_+_187740531 | 0.10 |
ENSRNOT00000092653
|
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr18_+_25163561 | 0.10 |
ENSRNOT00000042287
ENSRNOT00000049898 ENSRNOT00000017573 |
Bin1
|
bridging integrator 1 |
| chr1_-_107385257 | 0.09 |
ENSRNOT00000074296
|
Ccdc179
|
coiled-coil domain containing 179 |
| chr11_-_74866451 | 0.08 |
ENSRNOT00000002330
|
Il13ra1
|
interleukin 13 receptor subunit alpha 1 |
| chrX_+_15049462 | 0.08 |
ENSRNOT00000007015
|
Ebp
|
emopamil binding protein (sterol isomerase) |
| chr11_+_87374690 | 0.08 |
ENSRNOT00000086117
|
Aifm3
|
apoptosis inducing factor, mitochondria associated 3 |
| chr9_+_9842585 | 0.08 |
ENSRNOT00000070806
|
Cd70
|
Cd70 molecule |
| chr2_+_174203693 | 0.08 |
ENSRNOT00000073540
|
LOC108350036
|
uncharacterized LOC108350036 |
| chr3_-_176465162 | 0.08 |
ENSRNOT00000048807
|
Nkain4
|
Sodium/potassium transporting ATPase interacting 4 |
| chr1_+_216191886 | 0.07 |
ENSRNOT00000054863
|
Tspan32
|
tetraspanin 32 |
| chr13_-_109997092 | 0.07 |
ENSRNOT00000005190
|
Nenf
|
neudesin neurotrophic factor |
| chr13_-_99101208 | 0.07 |
ENSRNOT00000004329
|
H3f3a
|
H3 histone family member 3A |
| chr11_+_70056624 | 0.07 |
ENSRNOT00000002447
|
AC133403.1
|
|
| chr17_+_53956203 | 0.07 |
ENSRNOT00000022483
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr5_-_126221916 | 0.07 |
ENSRNOT00000052130
|
Lexm
|
lymphocyte expansion molecule |
| chr1_+_94718402 | 0.07 |
ENSRNOT00000046035
|
Uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr3_-_102826379 | 0.06 |
ENSRNOT00000073423
|
Olr769
|
olfactory receptor 769 |
| chr10_-_48038647 | 0.06 |
ENSRNOT00000078448
|
Prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chrX_-_14220662 | 0.06 |
ENSRNOT00000045753
|
Srpx
|
sushi-repeat-containing protein, X-linked |
| chr9_-_16612136 | 0.06 |
ENSRNOT00000023495
|
Mea1
|
male-enhanced antigen 1 |
| chr16_+_21275311 | 0.06 |
ENSRNOT00000027980
|
LOC100911483
|
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-like |
| chr17_+_23116661 | 0.06 |
ENSRNOT00000067374
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr14_-_60964324 | 0.06 |
ENSRNOT00000005155
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr1_+_100939740 | 0.06 |
ENSRNOT00000049584
|
Tsks
|
testis-specific serine kinase substrate |
| chr5_-_155258392 | 0.06 |
ENSRNOT00000017065
|
C1qc
|
complement C1q C chain |
| chr1_-_48825364 | 0.05 |
ENSRNOT00000024213
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr14_-_106393670 | 0.05 |
ENSRNOT00000011429
|
Mdh1
|
malate dehydrogenase 1 |
| chr6_-_127653124 | 0.05 |
ENSRNOT00000047324
|
Serpina11
|
serpin family A member 11 |
| chr7_-_29233392 | 0.05 |
ENSRNOT00000064241
|
Spic
|
Spi-C transcription factor |
| chr12_+_41463922 | 0.05 |
ENSRNOT00000089378
|
Cfap73
|
cilia and flagella associated protein 73 |
| chr9_-_15410943 | 0.05 |
ENSRNOT00000074217
|
Ccnd3
|
cyclin D3 |
| chr3_+_95233874 | 0.04 |
ENSRNOT00000079743
|
LOC691083
|
hypothetical protein LOC691083 |
| chrX_+_28593405 | 0.04 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chr1_+_101012822 | 0.04 |
ENSRNOT00000027809
|
Rras
|
related RAS viral (r-ras) oncogene homolog |
| chr6_-_102372611 | 0.04 |
ENSRNOT00000085927
|
Rdh11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr15_-_12889102 | 0.04 |
ENSRNOT00000061214
ENSRNOT00000084687 |
RGD1306063
|
similar to HT021 |
| chr18_-_52017734 | 0.04 |
ENSRNOT00000081020
|
March3
|
membrane associated ring-CH-type finger 3 |
| chr3_-_15376767 | 0.04 |
ENSRNOT00000044885
|
Ndufa8
|
NADH:ubiquinone oxidoreductase subunit A8 |
| chr20_+_1749716 | 0.03 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr16_-_2048292 | 0.03 |
ENSRNOT00000061883
|
Zcchc24
|
zinc finger CCHC-type containing 24 |
| chrX_-_1718637 | 0.03 |
ENSRNOT00000064309
|
Cdk16
|
cyclin-dependent kinase 16 |
| chr16_+_81089292 | 0.03 |
ENSRNOT00000087192
ENSRNOT00000026150 |
Tfdp1
|
transcription factor Dp-1 |
| chrM_+_5323 | 0.03 |
ENSRNOT00000050156
|
Mt-co1
|
mitochondrially encoded cytochrome c oxidase 1 |
| chr2_-_178612470 | 0.03 |
ENSRNOT00000013499
|
Tmem144
|
transmembrane protein 144 |
| chr1_+_227892956 | 0.03 |
ENSRNOT00000028483
|
AABR07006278.1
|
|
| chr8_+_85355766 | 0.03 |
ENSRNOT00000010583
|
Gcm1
|
glial cells missing homolog 1 |
| chr1_+_89202527 | 0.03 |
ENSRNOT00000028526
|
Sbsn
|
suprabasin |
| chr7_-_29282039 | 0.03 |
ENSRNOT00000067777
|
RGD1561102
|
similar to ribosomal protein S12 |
| chr6_+_135866739 | 0.03 |
ENSRNOT00000013460
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr20_+_10123651 | 0.03 |
ENSRNOT00000001559
|
Pde9a
|
phosphodiesterase 9A |
| chr2_-_27287605 | 0.03 |
ENSRNOT00000034041
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chrX_+_124894466 | 0.03 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr19_+_45946236 | 0.02 |
ENSRNOT00000045250
|
Syce1l
|
synaptonemal complex central element protein 1-like |
| chr8_-_79715284 | 0.02 |
ENSRNOT00000088562
|
Pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr5_+_28395296 | 0.02 |
ENSRNOT00000009375
|
Tmem55a
|
transmembrane protein 55A |
| chr1_+_167944448 | 0.02 |
ENSRNOT00000087667
|
Olr56
|
olfactory receptor 56 |
| chrX_+_105537602 | 0.02 |
ENSRNOT00000029833
|
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr1_+_84304228 | 0.02 |
ENSRNOT00000024771
|
Prx
|
periaxin |
| chr1_-_82279145 | 0.02 |
ENSRNOT00000057433
|
Cxcl17
|
C-X-C motif chemokine ligand 17 |
| chr16_-_21274688 | 0.02 |
ENSRNOT00000027953
|
Tssk6
|
testis-specific serine kinase 6 |
| chr5_+_104362971 | 0.02 |
ENSRNOT00000058520
|
Adamtsl1
|
ADAMTS-like 1 |
| chr15_-_33656089 | 0.02 |
ENSRNOT00000024186
|
Myh7
|
myosin heavy chain 7 |
| chr1_-_164307084 | 0.02 |
ENSRNOT00000086091
|
Serpinh1
|
serpin family H member 1 |
| chr9_+_12633990 | 0.02 |
ENSRNOT00000066517
ENSRNOT00000077532 |
Dazl
|
deleted in azoospermia-like |
| chr5_+_144308611 | 0.02 |
ENSRNOT00000014388
|
Col8a2
|
collagen type VIII alpha 2 chain |
| chr4_+_88584242 | 0.02 |
ENSRNOT00000008973
|
Pyurf
|
PIGY upstream reading frame |
| chrX_-_30831483 | 0.02 |
ENSRNOT00000004443
|
Glra2
|
glycine receptor, alpha 2 |
| chr5_-_144779212 | 0.02 |
ENSRNOT00000016230
|
Ncdn
|
neurochondrin |
| chr12_-_52124779 | 0.02 |
ENSRNOT00000088839
|
Galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr4_+_175431904 | 0.02 |
ENSRNOT00000032843
|
Pde3a
|
phosphodiesterase 3A |
| chr1_+_22364551 | 0.02 |
ENSRNOT00000043157
|
LOC108348200
|
trace amine-associated receptor 8a |
| chr4_-_70659252 | 0.02 |
ENSRNOT00000048049
|
Try10
|
trypsin 10 |
| chr7_+_107467260 | 0.01 |
ENSRNOT00000009240
ENSRNOT00000077382 |
Tg
|
thyroglobulin |
| chr14_+_106393959 | 0.01 |
ENSRNOT00000092168
|
Wdpcp
|
WD repeat containing planar cell polarity effector |
| chr1_-_100537377 | 0.01 |
ENSRNOT00000026599
|
Spib
|
Spi-B transcription factor |
| chr7_+_1206648 | 0.01 |
ENSRNOT00000073689
|
Pros1
|
protein S (alpha) |
| chr12_+_37702404 | 0.01 |
ENSRNOT00000049985
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr9_+_16612433 | 0.01 |
ENSRNOT00000023979
|
Klhdc3
|
kelch domain containing 3 |
| chr20_+_12052693 | 0.01 |
ENSRNOT00000083851
|
Adarb1
|
adenosine deaminase, RNA-specific, B1 |
| chr11_-_69223158 | 0.01 |
ENSRNOT00000067060
|
Mylk
|
myosin light chain kinase |
| chr12_-_31629881 | 0.01 |
ENSRNOT00000001238
|
Piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr19_+_56136904 | 0.01 |
ENSRNOT00000022823
|
Spire2
|
spire-type actin nucleation factor 2 |
| chr10_+_103349172 | 0.01 |
ENSRNOT00000035963
|
Gpr142
|
G protein-coupled receptor 142 |
| chr4_+_180291389 | 0.01 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr14_+_85113578 | 0.01 |
ENSRNOT00000011158
|
Nipsnap1
|
nipsnap homolog 1 |
| chr16_-_69961162 | 0.01 |
ENSRNOT00000074955
|
Prrg3
|
proline rich and Gla domain 3 |
| chr11_+_58624198 | 0.01 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chr5_+_144779681 | 0.01 |
ENSRNOT00000016493
|
LOC100294508
|
dyslexia susceptibility 2-like |
| chr1_-_82580007 | 0.01 |
ENSRNOT00000078668
|
Axl
|
Axl receptor tyrosine kinase |
| chr10_-_45480999 | 0.00 |
ENSRNOT00000078353
ENSRNOT00000084697 ENSRNOT00000087926 |
Obscn
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr15_+_42653148 | 0.00 |
ENSRNOT00000022095
|
Clu
|
clusterin |
| chr5_-_28349927 | 0.00 |
ENSRNOT00000008725
|
Otud6b
|
OTU domain containing 6B |
| chr9_+_38544398 | 0.00 |
ENSRNOT00000016828
|
Prim2
|
primase (DNA) subunit 2 |
| chr1_-_256914260 | 0.00 |
ENSRNOT00000020620
|
Fra10ac1
|
FRA10A associated CGG repeat 1 |
| chr3_-_4405194 | 0.00 |
ENSRNOT00000072025
|
AABR07051266.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0097374 | proprioception(GO:0019230) sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:0035772 | interleukin-13-mediated signaling pathway(GO:0035772) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |