Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
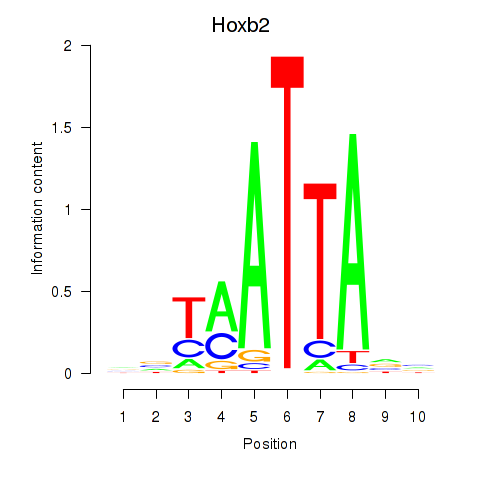
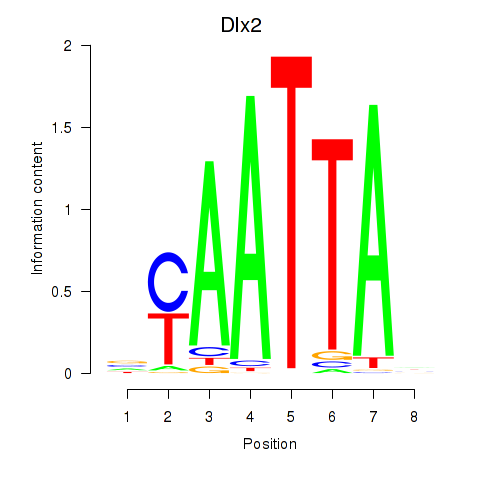
Results for Hoxb2_Dlx2
Z-value: 0.51
Transcription factors associated with Hoxb2_Dlx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb2
|
ENSRNOG00000008365 | homeobox B2 |
|
Dlx2
|
ENSRNOG00000001519 | distal-less homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb2 | rn6_v1_chr10_+_84200880_84200880 | 0.80 | 1.1e-01 | Click! |
| Dlx2 | rn6_v1_chr3_-_58181971_58181971 | 0.12 | 8.5e-01 | Click! |
Activity profile of Hoxb2_Dlx2 motif
Sorted Z-values of Hoxb2_Dlx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_2174131 | 1.87 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr3_-_66417741 | 1.09 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr10_-_88670430 | 0.42 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr12_-_17186679 | 0.41 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr2_+_145174876 | 0.40 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr4_+_94696965 | 0.36 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr17_+_11683862 | 0.30 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr6_-_125723732 | 0.22 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr2_-_185852759 | 0.22 |
ENSRNOT00000049461
|
Mab21l2
|
mab-21 like 2 |
| chr3_+_95715193 | 0.21 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr1_+_87224677 | 0.20 |
ENSRNOT00000028070
|
Ppp1r14a
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr6_-_106971250 | 0.19 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr12_-_45801842 | 0.19 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr1_+_242572533 | 0.17 |
ENSRNOT00000035123
|
Tmem252
|
transmembrane protein 252 |
| chr4_-_82173207 | 0.16 |
ENSRNOT00000074167
|
Hoxa5
|
homeo box A5 |
| chr16_+_54332660 | 0.15 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chrX_-_40086870 | 0.15 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr1_-_124803363 | 0.14 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr4_-_80395502 | 0.13 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr4_+_68656928 | 0.12 |
ENSRNOT00000016232
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr15_-_30147793 | 0.12 |
ENSRNOT00000060399
|
AABR07017693.1
|
|
| chr10_-_87286387 | 0.12 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr19_-_37528011 | 0.11 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr1_-_90149991 | 0.11 |
ENSRNOT00000076987
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr3_+_14889510 | 0.11 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr6_+_58468155 | 0.10 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr3_+_56766475 | 0.10 |
ENSRNOT00000078819
|
Sp5
|
Sp5 transcription factor |
| chr10_-_91986632 | 0.10 |
ENSRNOT00000087824
|
Nsf
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr8_+_71914867 | 0.10 |
ENSRNOT00000023372
|
Dapk2
|
death-associated protein kinase 2 |
| chr5_-_150167077 | 0.10 |
ENSRNOT00000088493
ENSRNOT00000013761 ENSRNOT00000067349 ENSRNOT00000051097 ENSRNOT00000067189 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chrX_+_118197217 | 0.09 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr2_-_33025271 | 0.09 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr18_-_32670665 | 0.09 |
ENSRNOT00000019409
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr15_+_44441856 | 0.09 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr4_+_117962319 | 0.09 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr1_+_105284753 | 0.09 |
ENSRNOT00000041950
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr8_-_78233430 | 0.09 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr5_-_12526962 | 0.08 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr6_-_125723944 | 0.08 |
ENSRNOT00000007004
|
Fbln5
|
fibulin 5 |
| chr3_+_45683993 | 0.08 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr10_-_87232723 | 0.08 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr4_+_158088505 | 0.07 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr10_-_93675991 | 0.07 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr7_+_72924799 | 0.07 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr19_+_25043680 | 0.07 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr12_-_10335499 | 0.07 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chr6_+_28382962 | 0.07 |
ENSRNOT00000016976
|
Pomc
|
proopiomelanocortin |
| chr2_-_235249571 | 0.07 |
ENSRNOT00000093631
ENSRNOT00000085096 |
Rrh
|
retinal pigment epithelium derived rhodopsin homolog |
| chr4_-_180234804 | 0.07 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr3_-_161917285 | 0.07 |
ENSRNOT00000025150
|
Cdh22
|
cadherin 22 |
| chr2_+_226563050 | 0.06 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr1_+_13595295 | 0.06 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr12_+_17253791 | 0.06 |
ENSRNOT00000083814
|
Zfand2a
|
zinc finger AN1-type containing 2A |
| chr1_-_215033460 | 0.06 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr3_+_128828331 | 0.06 |
ENSRNOT00000045393
|
Plcb4
|
phospholipase C, beta 4 |
| chr4_+_154423209 | 0.06 |
ENSRNOT00000075799
|
LOC100911545
|
alpha-2-macroglobulin-like |
| chr3_+_151335292 | 0.06 |
ENSRNOT00000073642
|
Mmp24
|
matrix metallopeptidase 24 |
| chr18_-_26656879 | 0.06 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr9_+_73418607 | 0.06 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chrX_+_33895769 | 0.06 |
ENSRNOT00000085993
ENSRNOT00000079365 |
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr12_+_24978483 | 0.06 |
ENSRNOT00000040069
|
Eln
|
elastin |
| chr4_+_172119331 | 0.06 |
ENSRNOT00000010579
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr1_-_48825364 | 0.05 |
ENSRNOT00000024213
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chrX_+_111735820 | 0.05 |
ENSRNOT00000086948
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chrX_-_124464963 | 0.05 |
ENSRNOT00000036472
ENSRNOT00000077697 |
Tmem255a
|
transmembrane protein 255A |
| chr5_-_136965191 | 0.05 |
ENSRNOT00000056842
|
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr12_+_50407843 | 0.05 |
ENSRNOT00000073763
|
Cryba4
|
crystallin, beta A4 |
| chr17_-_2705123 | 0.05 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr3_-_148932878 | 0.05 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chr13_+_34400170 | 0.05 |
ENSRNOT00000061516
ENSRNOT00000061515 ENSRNOT00000061513 ENSRNOT00000084506 ENSRNOT00000086641 |
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr1_-_67065797 | 0.05 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr2_+_185846232 | 0.05 |
ENSRNOT00000023418
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr5_+_58855773 | 0.05 |
ENSRNOT00000072869
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr12_+_24761210 | 0.05 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr2_+_51672722 | 0.05 |
ENSRNOT00000016485
|
Fgf10
|
fibroblast growth factor 10 |
| chr15_-_95514259 | 0.05 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr6_+_101532518 | 0.05 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr18_+_30435119 | 0.04 |
ENSRNOT00000027190
|
Pcdhb8
|
protocadherin beta 8 |
| chr9_+_73493027 | 0.04 |
ENSRNOT00000074427
ENSRNOT00000089478 |
Unc80
|
unc-80 homolog, NALCN activator |
| chr15_+_11298478 | 0.04 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr18_+_30592794 | 0.04 |
ENSRNOT00000027133
|
Pcdhb22
|
protocadherin beta 22 |
| chr10_+_66942398 | 0.04 |
ENSRNOT00000018986
|
Rab11fip4
|
RAB11 family interacting protein 4 |
| chrX_-_104932508 | 0.04 |
ENSRNOT00000075325
|
Nox1
|
NADPH oxidase 1 |
| chr9_-_65736852 | 0.04 |
ENSRNOT00000087458
|
Trak2
|
trafficking kinesin protein 2 |
| chr1_+_204959174 | 0.04 |
ENSRNOT00000023257
|
Zranb1
|
zinc finger RANBP2-type containing 1 |
| chr7_+_42269784 | 0.04 |
ENSRNOT00000008471
ENSRNOT00000007231 |
Kitlg
|
KIT ligand |
| chr2_-_96509424 | 0.04 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr3_+_54253949 | 0.04 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr14_+_76866135 | 0.04 |
ENSRNOT00000037291
|
Zfp518b
|
zinc finger protein 518B |
| chr10_+_91217079 | 0.04 |
ENSRNOT00000004218
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr2_-_196377883 | 0.04 |
ENSRNOT00000028661
|
Gabpb2
|
GA binding protein transcription factor, beta subunit 2 |
| chr7_-_28711761 | 0.04 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr17_+_63635086 | 0.03 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr5_+_50381244 | 0.03 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr4_-_17594598 | 0.03 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr2_-_30634243 | 0.03 |
ENSRNOT00000077537
|
Marveld2
|
MARVEL domain containing 2 |
| chr18_-_6781841 | 0.03 |
ENSRNOT00000077606
ENSRNOT00000048109 |
Aqp4
|
aquaporin 4 |
| chr10_-_84920886 | 0.03 |
ENSRNOT00000068083
|
Sp2
|
Sp2 transcription factor |
| chr1_-_173764246 | 0.03 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr16_+_46731403 | 0.03 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr2_+_80269661 | 0.03 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chr14_-_19072677 | 0.03 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr5_-_150704117 | 0.03 |
ENSRNOT00000067455
|
Sesn2
|
sestrin 2 |
| chr2_+_225645568 | 0.03 |
ENSRNOT00000017878
|
Abca4
|
ATP binding cassette subfamily A member 4 |
| chr4_+_22898527 | 0.03 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr9_+_71915421 | 0.03 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr20_+_1749716 | 0.03 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr10_+_17579284 | 0.03 |
ENSRNOT00000079698
|
Fbxw11
|
F-box and WD repeat domain containing 11 |
| chr6_-_67084234 | 0.03 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr9_-_65737538 | 0.03 |
ENSRNOT00000014939
|
Trak2
|
trafficking kinesin protein 2 |
| chr1_-_189182306 | 0.03 |
ENSRNOT00000021249
|
Gp2
|
glycoprotein 2 |
| chr6_-_111417813 | 0.03 |
ENSRNOT00000016324
ENSRNOT00000085458 |
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr3_+_59981959 | 0.03 |
ENSRNOT00000030460
|
Sp9
|
Sp9 transcription factor |
| chr18_+_30474947 | 0.02 |
ENSRNOT00000027188
|
Pcdhb9
|
protocadherin beta 9 |
| chr20_+_3558827 | 0.02 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr8_+_129617812 | 0.02 |
ENSRNOT00000079085
|
Ctnnb1
|
catenin beta 1 |
| chr4_-_100252755 | 0.02 |
ENSRNOT00000017301
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chrX_+_43881246 | 0.02 |
ENSRNOT00000005153
|
LOC317456
|
hypothetical LOC317456 |
| chr3_-_57607683 | 0.02 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chrX_+_110789269 | 0.02 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr5_+_135574172 | 0.02 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr6_+_8886591 | 0.02 |
ENSRNOT00000091510
ENSRNOT00000089174 |
Six3
|
SIX homeobox 3 |
| chr7_-_59587382 | 0.02 |
ENSRNOT00000091427
ENSRNOT00000066396 |
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr9_-_85243001 | 0.02 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr18_+_30509393 | 0.02 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr10_-_74679858 | 0.02 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chrX_+_65040934 | 0.02 |
ENSRNOT00000044006
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr19_+_39229754 | 0.02 |
ENSRNOT00000050612
|
Vps4a
|
vacuolar protein sorting 4 homolog A |
| chr5_-_164714145 | 0.02 |
ENSRNOT00000055680
|
LOC100911485
|
mitofusin-2-like |
| chr1_-_134871167 | 0.02 |
ENSRNOT00000076300
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr5_-_151473750 | 0.02 |
ENSRNOT00000011357
|
Tmem222
|
transmembrane protein 222 |
| chr1_+_185210922 | 0.02 |
ENSRNOT00000055120
|
Pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr7_-_116936674 | 0.02 |
ENSRNOT00000029456
|
Eef1d
|
eukaryotic translation elongation factor 1 delta |
| chr18_-_31109025 | 0.02 |
ENSRNOT00000026381
|
Fchsd1
|
FCH and double SH3 domains 1 |
| chr12_-_52658275 | 0.02 |
ENSRNOT00000041981
|
Zfp605
|
zinc finger protein 605 |
| chr6_-_3355339 | 0.02 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr2_+_266315036 | 0.02 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr3_-_60765645 | 0.02 |
ENSRNOT00000050513
|
Atf2
|
activating transcription factor 2 |
| chr9_-_53315915 | 0.02 |
ENSRNOT00000038093
|
Mstn
|
myostatin |
| chr1_+_193076430 | 0.02 |
ENSRNOT00000086492
ENSRNOT00000017885 |
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr5_+_139394794 | 0.02 |
ENSRNOT00000045954
|
Scmh1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr15_-_29903755 | 0.02 |
ENSRNOT00000066802
|
AABR07017669.1
|
|
| chr5_+_122019301 | 0.02 |
ENSRNOT00000068158
|
Pde4b
|
phosphodiesterase 4B |
| chr3_-_37803112 | 0.02 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr10_+_19366793 | 0.02 |
ENSRNOT00000050610
|
Fam196b
|
family with sequence similarity 196, member B |
| chr18_-_31343797 | 0.02 |
ENSRNOT00000081850
|
Pcdh1
|
protocadherin 1 |
| chr14_-_21128505 | 0.01 |
ENSRNOT00000004776
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr1_+_8310577 | 0.01 |
ENSRNOT00000015131
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr14_-_72380330 | 0.01 |
ENSRNOT00000082653
ENSRNOT00000081878 ENSRNOT00000006727 |
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr9_-_18371856 | 0.01 |
ENSRNOT00000027293
|
Supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr16_-_25192675 | 0.01 |
ENSRNOT00000032289
|
March1
|
membrane associated ring-CH-type finger 1 |
| chr17_+_24416651 | 0.01 |
ENSRNOT00000024458
|
Cd83
|
CD83 molecule |
| chr15_+_31950986 | 0.01 |
ENSRNOT00000080233
|
AABR07017868.4
|
|
| chr7_-_101138860 | 0.01 |
ENSRNOT00000077137
|
AABR07058124.4
|
|
| chr5_+_151362019 | 0.01 |
ENSRNOT00000077083
ENSRNOT00000067801 |
Wasf2
|
WAS protein family, member 2 |
| chr3_+_48106099 | 0.01 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr6_-_1003905 | 0.01 |
ENSRNOT00000082071
ENSRNOT00000042735 |
Fez2
|
fasciculation and elongation protein zeta 2 |
| chr2_-_57935334 | 0.01 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr16_+_63958645 | 0.01 |
ENSRNOT00000081522
ENSRNOT00000082355 ENSRNOT00000013991 |
Nrg1
|
neuregulin 1 |
| chr4_+_61924013 | 0.01 |
ENSRNOT00000090717
|
Bpgm
|
bisphosphoglycerate mutase |
| chrX_+_124321551 | 0.01 |
ENSRNOT00000074486
|
LOC100910807
|
transcriptional regulator Kaiso-like |
| chr2_-_177924970 | 0.01 |
ENSRNOT00000029340
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr13_+_87986240 | 0.01 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr5_+_36566783 | 0.01 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr1_-_43638161 | 0.01 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr16_+_23553647 | 0.01 |
ENSRNOT00000041994
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr10_-_72196437 | 0.01 |
ENSRNOT00000029769
|
Pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr9_-_81566096 | 0.01 |
ENSRNOT00000019497
|
Aamp
|
angio-associated, migratory cell protein |
| chr3_+_148635775 | 0.01 |
ENSRNOT00000067261
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr7_-_107391184 | 0.01 |
ENSRNOT00000056793
|
Tmem71
|
transmembrane protein 71 |
| chr6_-_123577695 | 0.01 |
ENSRNOT00000006604
|
Foxn3
|
forkhead box N3 |
| chr6_+_73553210 | 0.01 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr19_+_53044379 | 0.01 |
ENSRNOT00000072369
|
Foxc2
|
forkhead box C2 |
| chr6_-_7058314 | 0.01 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr1_-_101819478 | 0.01 |
ENSRNOT00000056181
|
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr10_-_87459652 | 0.01 |
ENSRNOT00000018804
|
Krt40
|
keratin 40 |
| chrX_-_77559348 | 0.01 |
ENSRNOT00000047823
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr7_+_2459141 | 0.01 |
ENSRNOT00000075681
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr7_-_101138373 | 0.01 |
ENSRNOT00000043257
|
LOC500877
|
Ab1-152 |
| chr14_+_76732650 | 0.01 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr7_+_42304534 | 0.01 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr2_+_154604832 | 0.01 |
ENSRNOT00000013777
|
Vom2r44
|
vomeronasal 2 receptor 44 |
| chr8_-_104593625 | 0.01 |
ENSRNOT00000016625
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr9_+_81566074 | 0.01 |
ENSRNOT00000074131
ENSRNOT00000046229 ENSRNOT00000090383 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr5_-_28131133 | 0.01 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr18_+_79773608 | 0.01 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr16_-_74264142 | 0.01 |
ENSRNOT00000026067
|
Dkk4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr14_-_79464770 | 0.01 |
ENSRNOT00000008932
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr16_-_73152921 | 0.01 |
ENSRNOT00000048602
|
Zmat4
|
zinc finger, matrin type 4 |
| chr16_+_35934970 | 0.01 |
ENSRNOT00000084707
ENSRNOT00000016474 |
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr3_+_56125924 | 0.01 |
ENSRNOT00000011380
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr5_-_38923095 | 0.00 |
ENSRNOT00000009146
|
AABR07047593.1
|
|
| chr14_-_16903242 | 0.00 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr5_+_146656049 | 0.00 |
ENSRNOT00000036029
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr3_+_48096954 | 0.00 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr4_+_68608137 | 0.00 |
ENSRNOT00000036398
|
Wee2
|
WEE1 homolog 2 |
| chrX_+_69730242 | 0.00 |
ENSRNOT00000075980
ENSRNOT00000076425 |
Eda
|
ectodysplasin-A |
| chr4_-_67610710 | 0.00 |
ENSRNOT00000039067
|
Mrps33
|
mitochondrial ribosomal protein S33 |
| chrX_+_159158194 | 0.00 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr13_+_27465930 | 0.00 |
ENSRNOT00000003314
|
Serpinb10
|
serpin family B member 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb2_Dlx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.2 | 1.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.3 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.1 | 0.4 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.2 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.1 | 0.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.0 | 0.1 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.4 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.1 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.0 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.0 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.0 | GO:0070662 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.0 | 0.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.4 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.0 | 0.0 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |