Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
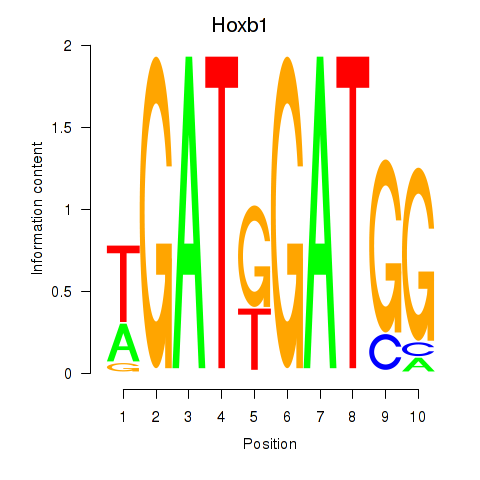
Results for Hoxb1
Z-value: 0.21
Transcription factors associated with Hoxb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb1
|
ENSRNOG00000008506 | homeo box B1 |
Activity profile of Hoxb1 motif
Sorted Z-values of Hoxb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_13810180 | 0.09 |
ENSRNOT00000084865
ENSRNOT00000038203 |
Tnrc18
|
trinucleotide repeat containing 18 |
| chr16_+_46731403 | 0.08 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr10_+_32824 | 0.07 |
ENSRNOT00000077828
|
LOC103689943
|
meiosis arrest female protein 1-like |
| chr1_-_267245636 | 0.07 |
ENSRNOT00000082799
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr1_-_260992291 | 0.07 |
ENSRNOT00000035415
ENSRNOT00000034758 |
Slit1
|
slit guidance ligand 1 |
| chr6_+_58468155 | 0.06 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr9_-_52238564 | 0.06 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr5_-_12526962 | 0.05 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr15_-_51168384 | 0.05 |
ENSRNOT00000021636
|
Slc25a37
|
solute carrier family 25 member 37 |
| chr3_+_116899878 | 0.05 |
ENSRNOT00000090802
ENSRNOT00000066101 |
Sema6d
|
semaphorin 6D |
| chr7_+_27089278 | 0.04 |
ENSRNOT00000082398
|
Nfyb
|
nuclear transcription factor Y subunit beta |
| chr6_+_52663112 | 0.04 |
ENSRNOT00000013842
|
Atxn7l1
|
ataxin 7-like 1 |
| chr11_-_38457373 | 0.04 |
ENSRNOT00000041177
|
Zfp295
|
zinc finger protein 295 |
| chr10_+_5930298 | 0.04 |
ENSRNOT00000044626
|
Grin2a
|
glutamate ionotropic receptor NMDA type subunit 2A |
| chr1_+_199323628 | 0.04 |
ENSRNOT00000036187
|
Zfp646
|
zinc finger protein 646 |
| chr14_+_3506339 | 0.03 |
ENSRNOT00000002867
|
Tgfbr3
|
transforming growth factor beta receptor 3 |
| chr10_-_70515936 | 0.03 |
ENSRNOT00000082684
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr1_+_25173242 | 0.03 |
ENSRNOT00000016518
|
Clvs2
|
clavesin 2 |
| chr5_+_122641885 | 0.03 |
ENSRNOT00000086765
|
Mier1
|
mesoderm induction early response 1, transcriptional regulator |
| chr5_+_124021366 | 0.03 |
ENSRNOT00000009977
|
Dab1
|
DAB1, reelin adaptor protein |
| chr15_-_33285779 | 0.03 |
ENSRNOT00000036150
|
Cdh24
|
cadherin 24 |
| chr13_+_71107465 | 0.03 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr12_-_37682964 | 0.03 |
ENSRNOT00000001421
|
RGD1563482
|
similar to hypothetical protein FLJ38663 |
| chr6_+_8218696 | 0.03 |
ENSRNOT00000083470
|
Ppm1b
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr4_-_155275161 | 0.02 |
ENSRNOT00000032690
|
Rimklb
|
ribosomal modification protein rimK-like family member B |
| chr4_+_65637407 | 0.02 |
ENSRNOT00000047954
|
Trim24
|
tripartite motif-containing 24 |
| chr18_-_77579969 | 0.02 |
ENSRNOT00000034896
|
Sall3
|
spalt-like transcription factor 3 |
| chr19_-_43290363 | 0.01 |
ENSRNOT00000024248
|
St3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr16_-_71203609 | 0.01 |
ENSRNOT00000088458
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr1_+_40816107 | 0.01 |
ENSRNOT00000060767
|
Akap12
|
A-kinase anchoring protein 12 |
| chr11_+_47061354 | 0.01 |
ENSRNOT00000039997
|
Pcnp
|
PEST proteolytic signal containing nuclear protein |
| chr7_+_124680294 | 0.01 |
ENSRNOT00000014720
|
Mpped1
|
metallophosphoesterase domain containing 1 |
| chr16_-_74864816 | 0.01 |
ENSRNOT00000017164
|
Alg11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr1_-_137359072 | 0.01 |
ENSRNOT00000014553
|
Klhl25
|
kelch-like family member 25 |
| chr6_+_26387877 | 0.00 |
ENSRNOT00000076105
|
Fndc4
|
|
| chr1_-_215838209 | 0.00 |
ENSRNOT00000050760
|
Igf2
|
insulin-like growth factor 2 |
| chr3_-_79892429 | 0.00 |
ENSRNOT00000016191
|
Slc39a13
|
solute carrier family 39 member 13 |
| chrX_+_82143789 | 0.00 |
ENSRNOT00000003724
|
Pou3f4
|
POU class 3 homeobox 4 |
| chr16_+_74865516 | 0.00 |
ENSRNOT00000058072
|
Atp7b
|
ATPase copper transporting beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.0 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.0 | GO:0060938 | transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0021812 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |