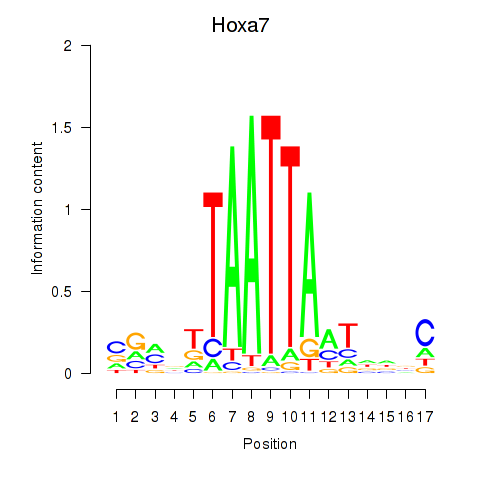
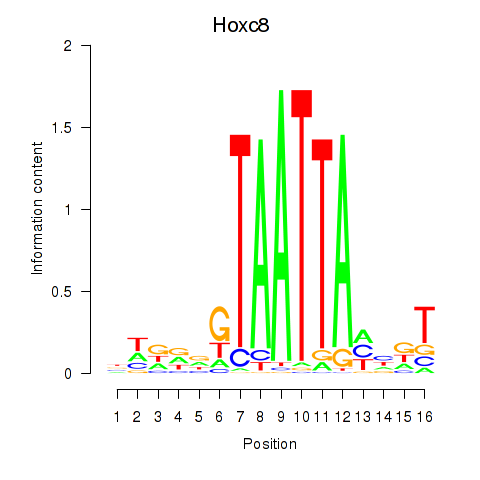
Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Hoxa7_Hoxc8
Z-value: 0.47
Transcription factors associated with Hoxa7_Hoxc8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa7
|
ENSRNOG00000049858 | homeobox A7 |
|
Hoxc8
|
ENSRNOG00000028619 | homeobox C8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa7 | rn6_v1_chr4_-_82271893_82271893 | 0.57 | 3.2e-01 | Click! |
| Hoxc8 | rn6_v1_chr7_+_144605058_144605058 | -0.47 | 4.3e-01 | Click! |
Activity profile of Hoxa7_Hoxc8 motif
Sorted Z-values of Hoxa7_Hoxc8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_14757679 | 0.22 |
ENSRNOT00000071364
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_+_141767940 | 0.21 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr2_-_33025271 | 0.14 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr10_+_61685645 | 0.13 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr6_+_64789940 | 0.13 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr5_+_25725683 | 0.13 |
ENSRNOT00000087602
|
LOC679087
|
similar to swan |
| chr15_+_62406873 | 0.13 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr1_+_99505677 | 0.12 |
ENSRNOT00000024645
|
Zfp719
|
zinc finger protein 719 |
| chr1_+_217345545 | 0.12 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr9_+_67234303 | 0.11 |
ENSRNOT00000050179
|
Abi2
|
abl-interactor 2 |
| chr1_-_43638161 | 0.11 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr15_-_30147793 | 0.11 |
ENSRNOT00000060399
|
AABR07017693.1
|
|
| chr8_-_128754514 | 0.11 |
ENSRNOT00000025019
|
Cx3cr1
|
C-X3-C motif chemokine receptor 1 |
| chr5_+_25253010 | 0.10 |
ENSRNOT00000061333
|
Rbm12b
|
RNA binding motif protein 12B |
| chr6_+_106052212 | 0.10 |
ENSRNOT00000010626
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr1_-_236729412 | 0.10 |
ENSRNOT00000054794
|
Prune2
|
prune homolog 2 |
| chr4_-_18396035 | 0.10 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr10_-_87286387 | 0.09 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr3_+_148654668 | 0.08 |
ENSRNOT00000081370
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr1_-_66212418 | 0.08 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr10_-_74679858 | 0.08 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr3_-_141411170 | 0.08 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr9_+_71915421 | 0.08 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr12_-_52658275 | 0.08 |
ENSRNOT00000041981
|
Zfp605
|
zinc finger protein 605 |
| chr12_-_45801842 | 0.07 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr17_+_11683862 | 0.07 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr9_+_73378057 | 0.07 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr11_-_61287914 | 0.07 |
ENSRNOT00000086286
|
Spice1
|
spindle and centriole associated protein 1 |
| chr8_+_71591360 | 0.07 |
ENSRNOT00000090096
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr8_-_69508024 | 0.07 |
ENSRNOT00000083564
|
AABR07070416.3
|
|
| chr15_-_55209342 | 0.06 |
ENSRNOT00000021752
|
Rb1
|
RB transcriptional corepressor 1 |
| chr2_+_72006099 | 0.06 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr3_+_48096954 | 0.06 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr5_-_133959447 | 0.06 |
ENSRNOT00000011985
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr8_+_33239139 | 0.06 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr10_-_62184874 | 0.06 |
ENSRNOT00000004229
|
Rpa1
|
replication protein A1 |
| chr3_-_15278645 | 0.06 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr2_+_145174876 | 0.06 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr9_+_84203072 | 0.06 |
ENSRNOT00000018882
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr4_+_94696965 | 0.05 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr9_+_73418607 | 0.05 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr4_+_148208260 | 0.05 |
ENSRNOT00000015742
|
Zfand4
|
zinc finger AN1-type containing 4 |
| chr10_-_46172166 | 0.05 |
ENSRNOT00000091052
ENSRNOT00000004412 |
Flcn
|
folliculin |
| chr4_+_157524423 | 0.05 |
ENSRNOT00000036654
|
Zfp384
|
zinc finger protein 384 |
| chr8_-_7426611 | 0.05 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr18_+_30880020 | 0.05 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr5_+_6373583 | 0.05 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr2_-_185852759 | 0.04 |
ENSRNOT00000049461
|
Mab21l2
|
mab-21 like 2 |
| chr14_+_39964588 | 0.04 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr8_-_39460844 | 0.04 |
ENSRNOT00000048875
|
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr1_-_241046249 | 0.04 |
ENSRNOT00000067796
ENSRNOT00000081866 |
Smc5
|
structural maintenance of chromosomes 5 |
| chr3_-_90751055 | 0.04 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr18_+_30820321 | 0.04 |
ENSRNOT00000060472
|
Pcdhga3
|
protocadherin gamma subfamily A, 3 |
| chr20_-_29184185 | 0.04 |
ENSRNOT00000090771
|
Mcu
|
mitochondrial calcium uniporter |
| chr4_-_150485781 | 0.04 |
ENSRNOT00000008763
|
Zfp248
|
zinc finger protein 248 |
| chr12_+_25430464 | 0.04 |
ENSRNOT00000002020
|
Gtf2i
|
general transcription factor II I |
| chrM_+_9870 | 0.03 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chrX_-_69218526 | 0.03 |
ENSRNOT00000092321
ENSRNOT00000074071 ENSRNOT00000092571 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chr5_+_58661049 | 0.03 |
ENSRNOT00000078274
|
Unc13b
|
unc-13 homolog B |
| chr1_+_101599018 | 0.03 |
ENSRNOT00000028494
|
Fut1
|
fucosyltransferase 1 |
| chr1_+_101603222 | 0.03 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr18_+_30592794 | 0.03 |
ENSRNOT00000027133
|
Pcdhb22
|
protocadherin beta 22 |
| chrX_+_84064427 | 0.03 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr2_-_35870578 | 0.03 |
ENSRNOT00000066069
|
Rnf180
|
ring finger protein 180 |
| chr3_-_72081079 | 0.03 |
ENSRNOT00000007914
|
Tmx2
|
thioredoxin-related transmembrane protein 2 |
| chr8_+_82038967 | 0.03 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr6_+_2216623 | 0.03 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr1_-_266428239 | 0.03 |
ENSRNOT00000027160
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr5_+_22392732 | 0.03 |
ENSRNOT00000087182
|
Clvs1
|
clavesin 1 |
| chr3_-_52447622 | 0.02 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr8_+_32188617 | 0.02 |
ENSRNOT00000081145
|
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr2_-_57935334 | 0.02 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr2_+_220432037 | 0.02 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr18_-_74059533 | 0.02 |
ENSRNOT00000038767
|
RGD1308601
|
similar to hypothetical protein |
| chr1_+_166141954 | 0.02 |
ENSRNOT00000042524
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr2_+_248398917 | 0.02 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr18_+_59748444 | 0.02 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr3_-_64095120 | 0.02 |
ENSRNOT00000016837
|
Sestd1
|
SEC14 and spectrin domain containing 1 |
| chr13_-_89545182 | 0.02 |
ENSRNOT00000078402
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr3_+_15560712 | 0.02 |
ENSRNOT00000010218
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr18_+_79773608 | 0.02 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr20_+_3155652 | 0.02 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr5_+_58995249 | 0.01 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr1_+_69841998 | 0.01 |
ENSRNOT00000072231
|
Zfp773-ps1
|
zinc finger protein 773, pseudogene 1 |
| chr3_-_160561741 | 0.01 |
ENSRNOT00000018364
|
Kcns1
|
potassium voltage-gated channel, modifier subfamily S, member 1 |
| chr6_+_44225233 | 0.01 |
ENSRNOT00000066593
|
Kidins220
|
kinase D-interacting substrate 220 |
| chr3_-_45169118 | 0.01 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr18_-_26656879 | 0.01 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chrX_-_36884748 | 0.01 |
ENSRNOT00000065678
|
Phka2
|
phosphorylase kinase, alpha 2 |
| chr1_+_255579474 | 0.01 |
ENSRNOT00000024465
|
Btaf1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr11_-_29710849 | 0.01 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr3_-_7498555 | 0.01 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr3_-_148057523 | 0.01 |
ENSRNOT00000055408
|
Defb24
|
defensin beta 24 |
| chr17_+_34704616 | 0.01 |
ENSRNOT00000090706
ENSRNOT00000083674 ENSRNOT00000077110 |
Exoc2
|
exocyst complex component 2 |
| chrX_+_9956370 | 0.01 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr11_-_82893845 | 0.01 |
ENSRNOT00000075306
|
LOC100912571
|
eukaryotic translation initiation factor 4 gamma 1-like |
| chrX_-_32095867 | 0.01 |
ENSRNOT00000049947
ENSRNOT00000080730 |
Ace2
|
angiotensin I converting enzyme 2 |
| chr10_+_76320041 | 0.01 |
ENSRNOT00000037693
|
Coil
|
coilin |
| chr1_+_201337416 | 0.01 |
ENSRNOT00000067742
|
Btbd16
|
BTB domain containing 16 |
| chr16_+_24978527 | 0.01 |
ENSRNOT00000019363
|
Tma16
|
translation machinery associated 16 homolog |
| chr8_+_36850324 | 0.01 |
ENSRNOT00000060448
|
Pate2
|
prostate and testis expressed 2 |
| chr1_-_23556241 | 0.00 |
ENSRNOT00000072943
|
LOC100910446
|
syntaxin-7-like |
| chr9_+_71247781 | 0.00 |
ENSRNOT00000049654
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr4_+_68608137 | 0.00 |
ENSRNOT00000036398
|
Wee2
|
WEE1 homolog 2 |
| chr10_-_71441389 | 0.00 |
ENSRNOT00000003699
|
Tada2a
|
transcriptional adaptor 2A |
| chr8_+_81863619 | 0.00 |
ENSRNOT00000080608
|
Fam214a
|
family with sequence similarity 214, member A |
| chr3_-_166994286 | 0.00 |
ENSRNOT00000081593
|
Zfp217
|
zinc finger protein 217 |
| chr10_+_95770154 | 0.00 |
ENSRNOT00000030300
|
Helz
|
helicase with zinc finger |
| chr16_+_50022998 | 0.00 |
ENSRNOT00000087986
|
Tlr3
|
toll-like receptor 3 |
| chr2_-_60657712 | 0.00 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr19_-_37528011 | 0.00 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr20_+_3830164 | 0.00 |
ENSRNOT00000045533
ENSRNOT00000084117 |
Col11a2
|
collagen type XI alpha 2 chain |
| chr2_+_196462554 | 0.00 |
ENSRNOT00000056380
|
Fam63a
|
family with sequence similarity 63, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa7_Hoxc8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0021888 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) regulation of post-translational protein modification(GO:1901873) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.0 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.0 | 0.0 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |