Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
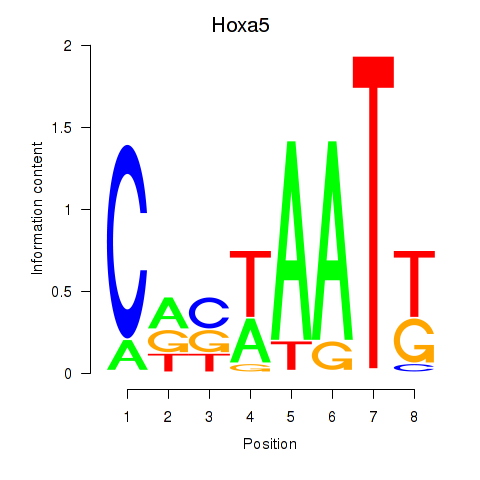
Results for Hoxa5
Z-value: 0.32
Transcription factors associated with Hoxa5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa5
|
ENSRNOG00000047951 | homeo box A5 |
|
Hoxa5
|
ENSRNOG00000006466 | homeo box A5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa5 | rn6_v1_chr4_-_82258765_82258765 | -0.54 | 3.5e-01 | Click! |
Activity profile of Hoxa5 motif
Sorted Z-values of Hoxa5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_44375804 | 0.13 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr1_-_37990007 | 0.09 |
ENSRNOT00000081054
|
AABR07001100.1
|
|
| chr1_-_101118825 | 0.09 |
ENSRNOT00000066328
|
Rps11
|
ribosomal protein S11 |
| chr11_+_86421106 | 0.09 |
ENSRNOT00000002599
|
LOC100362453
|
serine/threonine-protein phosphatase 2A catalytic subunit alpha-like |
| chr1_+_78417719 | 0.08 |
ENSRNOT00000020609
|
Tmem160
|
transmembrane protein 160 |
| chr5_-_17061361 | 0.08 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr9_-_30251388 | 0.08 |
ENSRNOT00000035033
|
Sdhaf4
|
succinate dehydrogenase complex assembly factor 4 |
| chr18_+_70739492 | 0.08 |
ENSRNOT00000085601
|
Acaa2
|
acetyl-CoA acyltransferase 2 |
| chr15_+_61879184 | 0.07 |
ENSRNOT00000042606
|
Sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr15_-_44442875 | 0.07 |
ENSRNOT00000017939
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr5_+_63192298 | 0.07 |
ENSRNOT00000008329
|
Sec61b
|
Sec61 translocon beta subunit |
| chrX_+_31984612 | 0.07 |
ENSRNOT00000005181
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr7_+_116357175 | 0.07 |
ENSRNOT00000076790
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr14_-_2032593 | 0.07 |
ENSRNOT00000000037
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr1_-_220746224 | 0.07 |
ENSRNOT00000027734
|
Banf1
|
barrier to autointegration factor 1 |
| chr10_-_56849255 | 0.07 |
ENSRNOT00000025491
|
RGD1308134
|
similar to RIKEN cDNA 1110020A23 |
| chr16_-_75530500 | 0.06 |
ENSRNOT00000018580
ENSRNOT00000086035 |
RatNP-3b
|
defensin RatNP-3 precursor |
| chr1_-_167698263 | 0.06 |
ENSRNOT00000093046
|
Trim21
|
tripartite motif-containing 21 |
| chr3_-_4341771 | 0.06 |
ENSRNOT00000034694
|
LOC684988
|
similar to ribosomal protein S13 |
| chr7_-_143967484 | 0.06 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr1_-_67094567 | 0.06 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr6_-_91581262 | 0.06 |
ENSRNOT00000034507
|
AABR07064716.1
|
|
| chr14_-_19132208 | 0.06 |
ENSRNOT00000060535
|
Afm
|
afamin |
| chr4_-_155923079 | 0.06 |
ENSRNOT00000013308
|
Clec4a3
|
C-type lectin domain family 4, member A3 |
| chr17_+_57033573 | 0.06 |
ENSRNOT00000020160
|
Crem
|
cAMP responsive element modulator |
| chrX_-_123980357 | 0.06 |
ENSRNOT00000049435
|
Rhox8
|
reproductive homeobox 8 |
| chr1_+_53220397 | 0.06 |
ENSRNOT00000089989
|
AABR07001573.1
|
|
| chr5_-_134927235 | 0.06 |
ENSRNOT00000016751
|
Uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr1_+_248647170 | 0.06 |
ENSRNOT00000015016
|
Tpd52l3
|
tumor protein D52-like 3 |
| chr20_-_10842370 | 0.06 |
ENSRNOT00000001580
|
Hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr4_+_165732643 | 0.06 |
ENSRNOT00000034403
|
LOC690326
|
hypothetical protein LOC690326 |
| chr9_+_68414339 | 0.06 |
ENSRNOT00000040778
|
Pard3b
|
par-3 family cell polarity regulator beta |
| chr3_-_150108898 | 0.06 |
ENSRNOT00000022914
|
Pxmp4
|
peroxisomal membrane protein 4 |
| chr7_-_144837395 | 0.06 |
ENSRNOT00000089024
|
Cbx5
|
chromobox 5 |
| chr9_+_44491177 | 0.06 |
ENSRNOT00000075242
|
NEWGENE_1308196
|
mitochondrial ribosomal protein L30 |
| chr11_+_89008008 | 0.06 |
ENSRNOT00000074586
|
Cebpd
|
CCAAT/enhancer binding protein delta |
| chr4_-_89151184 | 0.06 |
ENSRNOT00000010263
|
Nap1l5
|
nucleosome assembly protein 1-like 5 |
| chr10_-_66229311 | 0.06 |
ENSRNOT00000016897
|
Lgals5
|
lectin, galactose binding, soluble 5 |
| chr12_+_2170630 | 0.06 |
ENSRNOT00000071928
|
Pet100
|
PET100 homolog |
| chr7_-_144837583 | 0.05 |
ENSRNOT00000055289
|
Cbx5
|
chromobox 5 |
| chr2_-_235831670 | 0.05 |
ENSRNOT00000037393
|
Ostc
|
oligosaccharyltransferase complex non-catalytic subunit |
| chr6_+_52702544 | 0.05 |
ENSRNOT00000014252
|
Efcab10
|
EF-hand calcium binding domain 10 |
| chr7_-_145338152 | 0.05 |
ENSRNOT00000055268
|
Spt1
|
salivary protein 1 |
| chr3_-_8955538 | 0.05 |
ENSRNOT00000040570
|
LOC686066
|
similar to 60S ribosomal protein L38 |
| chr10_-_57671080 | 0.05 |
ENSRNOT00000082511
|
LOC691995
|
hypothetical protein LOC691995 |
| chr2_+_187447501 | 0.05 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr5_+_16845631 | 0.05 |
ENSRNOT00000047889
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr19_+_51985170 | 0.05 |
ENSRNOT00000019443
|
Hsbp1
|
heat shock factor binding protein 1 |
| chr13_-_32427177 | 0.05 |
ENSRNOT00000044628
|
Cdh19
|
cadherin 19 |
| chr9_-_113598477 | 0.05 |
ENSRNOT00000035606
ENSRNOT00000084884 |
Ralbp1
|
ralA binding protein 1 |
| chr10_+_95770154 | 0.05 |
ENSRNOT00000030300
|
Helz
|
helicase with zinc finger |
| chr7_-_144993652 | 0.05 |
ENSRNOT00000087748
|
Itga5
|
integrin subunit alpha 5 |
| chr10_-_89187474 | 0.05 |
ENSRNOT00000064931
|
Usmg5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr15_-_61873406 | 0.05 |
ENSRNOT00000045115
|
LOC100360244
|
LRRGT00053-like |
| chr14_+_22706901 | 0.05 |
ENSRNOT00000088798
|
Ppial4d
|
peptidylprolyl isomerase A (cyclophilin A)-like 4D |
| chrX_+_13441558 | 0.05 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr14_-_21709084 | 0.05 |
ENSRNOT00000087477
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chr6_+_72891725 | 0.05 |
ENSRNOT00000038074
|
Nubpl
|
nucleotide binding protein-like |
| chr11_+_45751812 | 0.05 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr20_+_3230052 | 0.05 |
ENSRNOT00000078454
|
RT1-T24-3
|
RT1 class I, locus T24, gene 3 |
| chr10_-_46404642 | 0.05 |
ENSRNOT00000083698
|
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr10_-_56850085 | 0.05 |
ENSRNOT00000025767
|
Rnasek
|
ribonuclease K |
| chr14_-_19159923 | 0.05 |
ENSRNOT00000003879
|
Afp
|
alpha-fetoprotein |
| chr2_-_198458041 | 0.05 |
ENSRNOT00000082450
|
Fcgr1a
|
Fc fragment of IgG receptor Ia |
| chr6_+_22296128 | 0.05 |
ENSRNOT00000087805
|
Dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr10_+_31647898 | 0.05 |
ENSRNOT00000066536
|
LOC689968
|
similar to kidney injury molecule 1 |
| chr5_+_135562034 | 0.05 |
ENSRNOT00000056967
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr1_-_166919302 | 0.05 |
ENSRNOT00000026925
|
Folr2
|
folate receptor beta |
| chr3_+_161121697 | 0.05 |
ENSRNOT00000036020
|
Wfdc10a
|
WAP four-disulfide core domain 10A |
| chr7_+_91384187 | 0.05 |
ENSRNOT00000005828
|
Utp23
|
UTP23, small subunit processome component |
| chr4_-_129430251 | 0.05 |
ENSRNOT00000067881
|
Fam19a4
|
family with sequence similarity 19 member A4, C-C motif chemokine like |
| chr11_-_71601662 | 0.05 |
ENSRNOT00000002397
|
Tctex1d2
|
Tctex1 domain containing 2 |
| chr4_-_162726628 | 0.05 |
ENSRNOT00000073877
|
LOC100911272
|
killer cell lectin-like receptor 5-like |
| chr7_-_143966863 | 0.05 |
ENSRNOT00000018828
|
Sp7
|
Sp7 transcription factor |
| chr5_+_2654065 | 0.05 |
ENSRNOT00000074957
|
LOC100912291
|
uncharacterized LOC100912291 |
| chr11_-_71523267 | 0.05 |
ENSRNOT00000033026
|
Zdhhc19
|
zinc finger, DHHC-type containing 19 |
| chr9_-_101388833 | 0.05 |
ENSRNOT00000071817
|
Chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr10_+_11146359 | 0.05 |
ENSRNOT00000006314
|
Pam16
|
presequence translocase associated motor 16 homolog |
| chr5_-_60541282 | 0.04 |
ENSRNOT00000093015
|
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr2_-_183128932 | 0.04 |
ENSRNOT00000031179
|
Mnd1
|
meiotic nuclear divisions 1 |
| chr11_+_53081025 | 0.04 |
ENSRNOT00000002700
|
Bbx
|
BBX, HMG-box containing |
| chr9_-_101107535 | 0.04 |
ENSRNOT00000080892
ENSRNOT00000071424 |
LOC100911516
|
coiled-coil-helix-coiled-coil-helix domain-containing protein 2, mitochondrial-like |
| chr17_+_69960160 | 0.04 |
ENSRNOT00000023887
|
Ucn3
|
urocortin 3 |
| chr17_+_44522140 | 0.04 |
ENSRNOT00000080490
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr2_+_150211898 | 0.04 |
ENSRNOT00000018767
|
Sucnr1
|
succinate receptor 1 |
| chr20_-_28873363 | 0.04 |
ENSRNOT00000071064
|
Snrpd2l
|
small nuclear ribonucleoprotein D2-like |
| chr8_+_21663325 | 0.04 |
ENSRNOT00000027749
|
Ubl5
|
ubiquitin-like 5 |
| chr14_+_59611434 | 0.04 |
ENSRNOT00000065366
|
Cckar
|
cholecystokinin A receptor |
| chr1_+_22364551 | 0.04 |
ENSRNOT00000043157
|
LOC108348200
|
trace amine-associated receptor 8a |
| chr1_-_37726151 | 0.04 |
ENSRNOT00000071842
|
RGD1308544
|
LOC361192 |
| chr4_-_164123974 | 0.04 |
ENSRNOT00000082447
|
LOC502907
|
similar to immunoreceptor Ly49si1 |
| chr9_-_10170428 | 0.04 |
ENSRNOT00000073048
|
LOC316124
|
similar to gonadotropin-regulated long chain acyl-CoA synthetase |
| chr4_+_152630469 | 0.04 |
ENSRNOT00000013721
|
Ninj2
|
ninjurin 2 |
| chr6_-_26138414 | 0.04 |
ENSRNOT00000034712
|
Mrpl33
|
mitochondrial ribosomal protein L33 |
| chr2_+_115739813 | 0.04 |
ENSRNOT00000085452
|
Slc7a14
|
solute carrier family 7, member 14 |
| chr9_-_121972055 | 0.04 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr1_+_1545134 | 0.04 |
ENSRNOT00000044523
|
AABR07000156.1
|
|
| chr2_-_164684985 | 0.04 |
ENSRNOT00000057504
|
Rarres1
|
retinoic acid receptor responder 1 |
| chr2_-_157066781 | 0.04 |
ENSRNOT00000001187
|
LOC304239
|
similar to RalA binding protein 1 |
| chr5_-_64818813 | 0.04 |
ENSRNOT00000009111
ENSRNOT00000086505 |
Aldob
|
aldolase, fructose-bisphosphate B |
| chr10_-_90312386 | 0.04 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr4_-_29778039 | 0.04 |
ENSRNOT00000074177
|
Sgce
|
sarcoglycan, epsilon |
| chrX_-_82699487 | 0.04 |
ENSRNOT00000081625
|
Rps6ka6
|
ribosomal protein S6 kinase A6 |
| chr3_+_80362858 | 0.04 |
ENSRNOT00000021353
ENSRNOT00000086242 |
Lrp4
|
LDL receptor related protein 4 |
| chr4_+_170958196 | 0.04 |
ENSRNOT00000007905
|
Pde6h
|
phosphodiesterase 6H |
| chr1_-_72335855 | 0.04 |
ENSRNOT00000021613
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr1_-_227359809 | 0.04 |
ENSRNOT00000088080
|
1700025F22Rik
|
RIKEN cDNA 1700025F22 gene |
| chr7_-_55604403 | 0.04 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr4_-_155401480 | 0.04 |
ENSRNOT00000020735
|
Apobec1
|
apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| chr7_+_140054592 | 0.04 |
ENSRNOT00000014337
|
Olr1108
|
olfactory receptor 1108 |
| chr20_-_45260119 | 0.04 |
ENSRNOT00000000718
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr1_-_263762785 | 0.04 |
ENSRNOT00000018221
|
Cpn1
|
carboxypeptidase N subunit 1 |
| chr1_+_282638017 | 0.04 |
ENSRNOT00000079722
ENSRNOT00000082254 |
Ces2c
|
carboxylesterase 2C |
| chr8_+_61615650 | 0.04 |
ENSRNOT00000007261
|
Snupn
|
snurportin 1 |
| chr6_+_109939345 | 0.04 |
ENSRNOT00000013560
|
Ift43
|
intraflagellar transport 43 |
| chr7_-_54778848 | 0.04 |
ENSRNOT00000005399
|
Glipr1
|
GLI pathogenesis-related 1 |
| chr17_-_43626538 | 0.04 |
ENSRNOT00000074292
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr9_+_20279938 | 0.04 |
ENSRNOT00000075612
|
Grcc10
|
gene rich cluster, C10 gene |
| chr20_+_2094931 | 0.04 |
ENSRNOT00000001013
|
Ppp1r11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr4_-_157266018 | 0.04 |
ENSRNOT00000019570
|
LOC100911713
|
protein C10-like |
| chr5_+_33097654 | 0.04 |
ENSRNOT00000008087
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr1_-_228014924 | 0.04 |
ENSRNOT00000042091
|
Oosp2
|
oocyte secreted protein 2 |
| chr3_+_66673071 | 0.04 |
ENSRNOT00000034769
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chrX_+_156355376 | 0.04 |
ENSRNOT00000078304
|
Lage3
|
L antigen family, member 3 |
| chr17_+_45175121 | 0.04 |
ENSRNOT00000080417
|
Nkapl
|
NFKB activating protein-like |
| chr9_+_18001105 | 0.04 |
ENSRNOT00000071814
|
AABR07066826.2
|
|
| chr20_-_1099336 | 0.04 |
ENSRNOT00000071766
|
LOC100362856
|
olfactory receptor-like protein-like |
| chrX_-_71169865 | 0.04 |
ENSRNOT00000050415
|
Il2rg
|
interleukin 2 receptor subunit gamma |
| chr10_+_63731767 | 0.04 |
ENSRNOT00000005368
|
Pitpna
|
phosphatidylinositol transfer protein, alpha |
| chr5_+_148051126 | 0.04 |
ENSRNOT00000074415
|
Ptp4a2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr19_+_14835822 | 0.04 |
ENSRNOT00000072804
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene |
| chr9_+_44567206 | 0.04 |
ENSRNOT00000088544
|
NEWGENE_1308196
|
mitochondrial ribosomal protein L30 |
| chr1_-_233140237 | 0.04 |
ENSRNOT00000083372
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr3_-_67668772 | 0.04 |
ENSRNOT00000010247
|
Frzb
|
frizzled-related protein |
| chr1_+_168945449 | 0.04 |
ENSRNOT00000087661
ENSRNOT00000019913 |
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr4_-_44136815 | 0.04 |
ENSRNOT00000086810
|
Tfec
|
transcription factor EC |
| chr15_+_12827707 | 0.04 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr4_-_98342887 | 0.04 |
ENSRNOT00000010156
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr18_+_83777665 | 0.04 |
ENSRNOT00000018682
|
Cbln2
|
cerebellin 2 precursor |
| chr16_+_73564243 | 0.04 |
ENSRNOT00000030960
|
Golga7
|
golgin A7 |
| chr13_+_49277722 | 0.04 |
ENSRNOT00000035335
|
Tmem81
|
transmembrane protein 81 |
| chr2_-_158156444 | 0.04 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr1_+_188278469 | 0.04 |
ENSRNOT00000065601
|
Tmc5
|
transmembrane channel-like 5 |
| chr9_-_99818262 | 0.04 |
ENSRNOT00000056600
|
Cops9
|
COP9 signalosome subunit 9 |
| chr4_-_82702429 | 0.04 |
ENSRNOT00000011069
|
Hibadh
|
3-hydroxyisobutyrate dehydrogenase |
| chr2_-_28799266 | 0.04 |
ENSRNOT00000089293
|
Tmem171
|
transmembrane protein 171 |
| chr1_+_224933920 | 0.04 |
ENSRNOT00000066823
|
Wdr74
|
WD repeat domain 74 |
| chr6_+_29977797 | 0.04 |
ENSRNOT00000071784
|
Fkbp1b
|
FK506 binding protein 1B |
| chr5_-_119564846 | 0.04 |
ENSRNOT00000012977
|
Cyp2j4
|
cytochrome P450, family 2, subfamily j, polypeptide 4 |
| chr18_+_55505993 | 0.04 |
ENSRNOT00000043736
|
RGD1309362
|
similar to interferon-inducible GTPase |
| chr3_-_127500709 | 0.03 |
ENSRNOT00000006330
|
Hao1
|
hydroxyacid oxidase 1 |
| chr2_-_140464607 | 0.03 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr1_-_206394346 | 0.03 |
ENSRNOT00000038122
|
LOC100302465
|
hypothetical LOC100302465 |
| chrX_-_142248369 | 0.03 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr20_+_5040337 | 0.03 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chrX_+_111396995 | 0.03 |
ENSRNOT00000087634
|
Pih1d3
|
PIH1 domain containing 3 |
| chr16_+_3817757 | 0.03 |
ENSRNOT00000088566
|
Tmem254
|
transmembrane protein 254 |
| chr2_-_122756842 | 0.03 |
ENSRNOT00000080181
|
Ccdc144b
|
coiled-coil domain containing 144B |
| chr19_+_40855433 | 0.03 |
ENSRNOT00000023823
|
Il34
|
interleukin 34 |
| chr3_-_102826379 | 0.03 |
ENSRNOT00000073423
|
Olr769
|
olfactory receptor 769 |
| chr4_-_13878126 | 0.03 |
ENSRNOT00000007032
|
Gnat3
|
G protein subunit alpha transducin 3 |
| chr20_+_4087618 | 0.03 |
ENSRNOT00000000522
ENSRNOT00000060327 ENSRNOT00000080590 |
RT1-Db1
|
RT1 class II, locus Db1 |
| chr5_-_63192025 | 0.03 |
ENSRNOT00000008913
|
Alg2
|
ALG2, alpha-1,3/1,6-mannosyltransferase |
| chr7_+_116357783 | 0.03 |
ENSRNOT00000076279
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr1_-_178367828 | 0.03 |
ENSRNOT00000050823
|
AABR07071968.1
|
|
| chr1_-_195096694 | 0.03 |
ENSRNOT00000088874
|
Snurf
|
SNRPN upstream reading frame |
| chr13_-_85622314 | 0.03 |
ENSRNOT00000005719
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr4_+_33890349 | 0.03 |
ENSRNOT00000078680
|
C1galt1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr15_+_12577440 | 0.03 |
ENSRNOT00000079000
|
Thoc7
|
THO complex 7 |
| chr1_-_155955173 | 0.03 |
ENSRNOT00000079345
|
AABR07004776.1
|
|
| chrX_+_28593405 | 0.03 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chr13_-_36101411 | 0.03 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr15_-_12129220 | 0.03 |
ENSRNOT00000029318
|
Olr1605
|
olfactory receptor 1605 |
| chrX_-_77418525 | 0.03 |
ENSRNOT00000092303
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr9_-_50762082 | 0.03 |
ENSRNOT00000015492
|
Mettl21c
|
methyltransferase like 21C |
| chr2_-_183250928 | 0.03 |
ENSRNOT00000081080
|
AABR07012039.1
|
|
| chr5_-_172307431 | 0.03 |
ENSRNOT00000018453
|
Fam213b
|
family with sequence similarity 213, member B |
| chr16_+_62153792 | 0.03 |
ENSRNOT00000074543
|
Smim18
|
small integral membrane protein 18 |
| chr1_-_195096460 | 0.03 |
ENSRNOT00000077253
|
Snrpn
|
small nuclear ribonucleoprotein polypeptide N |
| chr2_+_154604832 | 0.03 |
ENSRNOT00000013777
|
Vom2r44
|
vomeronasal 2 receptor 44 |
| chr9_-_19880346 | 0.03 |
ENSRNOT00000014051
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr9_-_27452902 | 0.03 |
ENSRNOT00000018325
|
Gsta1
|
glutathione S-transferase alpha 1 |
| chr2_-_188745144 | 0.03 |
ENSRNOT00000055533
|
Cks1b
|
CDC28 protein kinase regulatory subunit 1B |
| chr9_+_67130259 | 0.03 |
ENSRNOT00000023796
|
Cyp20a1
|
cytochrome P450, family 20, subfamily a, polypeptide 1 |
| chr8_-_14156656 | 0.03 |
ENSRNOT00000089364
|
Deup1
|
deuterosome assembly protein 1 |
| chr18_+_41022552 | 0.03 |
ENSRNOT00000005260
|
Commd10
|
COMM domain containing 10 |
| chr7_+_136037230 | 0.03 |
ENSRNOT00000071423
|
AABR07058745.1
|
|
| chr1_-_81550598 | 0.03 |
ENSRNOT00000051671
|
RGD1564380
|
similar to BC049730 protein |
| chr16_-_20869555 | 0.03 |
ENSRNOT00000092425
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr1_+_214183724 | 0.03 |
ENSRNOT00000091150
|
Lrrc56
|
leucine rich repeat containing 56 |
| chr14_+_17534412 | 0.03 |
ENSRNOT00000079304
ENSRNOT00000046771 |
Cdkl2
|
cyclin dependent kinase like 2 |
| chr5_-_102743417 | 0.03 |
ENSRNOT00000067389
|
Bnc2
|
basonuclin 2 |
| chr15_+_27875911 | 0.03 |
ENSRNOT00000013582
|
Pnp
|
purine nucleoside phosphorylase |
| chr1_-_64030175 | 0.03 |
ENSRNOT00000089950
|
Tsen34l1
|
tRNA splicing endonuclease subunit 34-like 1 |
| chr10_-_90127600 | 0.03 |
ENSRNOT00000028368
|
Lsm12
|
LSM12 homolog |
| chr10_+_89174684 | 0.03 |
ENSRNOT00000043754
|
Vps25
|
vacuolar protein sorting 25 |
| chr10_+_59894340 | 0.03 |
ENSRNOT00000080446
|
Spata22
|
spermatogenesis associated 22 |
| chr3_+_110855000 | 0.03 |
ENSRNOT00000081613
|
Knl1
|
kinetochore scaffold 1 |
| chr3_+_72080630 | 0.03 |
ENSRNOT00000008911
|
Med19
|
mediator complex subunit 19 |
| chr13_-_82005741 | 0.03 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.0 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.0 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.0 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.0 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.0 | GO:0070637 | nicotinamide riboside catabolic process(GO:0006738) urate biosynthetic process(GO:0034418) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.0 | 0.0 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.0 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.0 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.0 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.0 | GO:0047619 | acylcarnitine hydrolase activity(GO:0047619) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.0 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.0 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |