Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
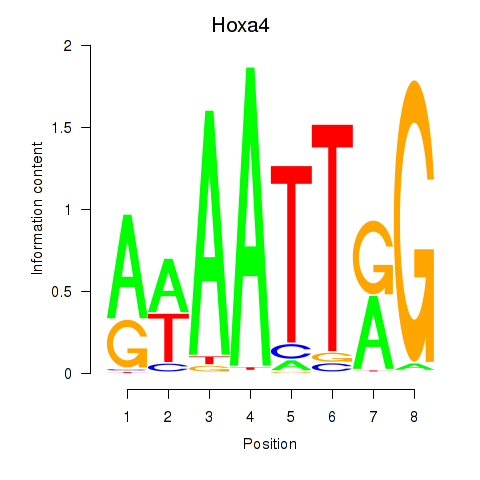
Results for Hoxa4
Z-value: 0.52
Transcription factors associated with Hoxa4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa4
|
ENSRNOG00000027365 | homeo box A4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa4 | rn6_v1_chr4_-_82160240_82160240 | 0.81 | 9.9e-02 | Click! |
Activity profile of Hoxa4 motif
Sorted Z-values of Hoxa4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_17186679 | 0.84 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr3_-_66417741 | 0.37 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr9_+_25410669 | 0.27 |
ENSRNOT00000030912
ENSRNOT00000090920 |
Tfap2b
|
transcription factor AP-2 beta |
| chr7_+_144647587 | 0.22 |
ENSRNOT00000022398
|
Hoxc4
|
homeo box C4 |
| chr2_+_206342066 | 0.18 |
ENSRNOT00000026556
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr5_-_12526962 | 0.17 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chrX_-_124464963 | 0.16 |
ENSRNOT00000036472
ENSRNOT00000077697 |
Tmem255a
|
transmembrane protein 255A |
| chr3_+_95715193 | 0.14 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chrX_+_158835811 | 0.12 |
ENSRNOT00000071888
ENSRNOT00000080110 |
Ints6l
|
integrator complex subunit 6 like |
| chr16_+_54332660 | 0.11 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr3_+_56766475 | 0.11 |
ENSRNOT00000078819
|
Sp5
|
Sp5 transcription factor |
| chrX_-_63165759 | 0.11 |
ENSRNOT00000076765
|
Zfx
|
zinc finger protein X-linked |
| chr9_+_121831716 | 0.11 |
ENSRNOT00000056250
ENSRNOT00000056251 |
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr1_-_163554839 | 0.10 |
ENSRNOT00000081509
|
Emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr4_-_82160240 | 0.10 |
ENSRNOT00000038775
ENSRNOT00000058985 |
Hoxa4
|
homeo box A4 |
| chr2_+_207930796 | 0.10 |
ENSRNOT00000047827
|
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr4_+_100166863 | 0.10 |
ENSRNOT00000014505
|
Sftpb
|
surfactant protein B |
| chr16_+_50152008 | 0.09 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr19_-_26053762 | 0.08 |
ENSRNOT00000004646
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr14_-_13058172 | 0.08 |
ENSRNOT00000002746
ENSRNOT00000071706 |
Prdm8
|
PR/SET domain 8 |
| chr8_+_110982777 | 0.08 |
ENSRNOT00000010992
|
Ky
|
kyphoscoliosis peptidase |
| chr10_-_87286387 | 0.08 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr6_-_51018050 | 0.07 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr6_+_96834525 | 0.07 |
ENSRNOT00000077935
|
Hif1a
|
hypoxia inducible factor 1 alpha subunit |
| chr1_-_33275540 | 0.07 |
ENSRNOT00000017019
|
Irx2
|
iroquois homeobox 2 |
| chr16_-_74700815 | 0.07 |
ENSRNOT00000081297
|
Thsd1
|
thrombospondin type 1 domain containing 1 |
| chr17_+_70262363 | 0.07 |
ENSRNOT00000048933
|
Fam208b
|
family with sequence similarity 208, member B |
| chr1_+_140998240 | 0.07 |
ENSRNOT00000023506
ENSRNOT00000090897 |
Abhd2
|
abhydrolase domain containing 2 |
| chr3_-_80601410 | 0.07 |
ENSRNOT00000022841
|
Atg13
|
autophagy related 13 |
| chr3_-_146396299 | 0.07 |
ENSRNOT00000040188
ENSRNOT00000008931 |
Apmap
|
adipocyte plasma membrane associated protein |
| chrX_-_135419704 | 0.06 |
ENSRNOT00000044435
|
Zfp280c
|
zinc finger protein 280C |
| chr7_-_140172448 | 0.06 |
ENSRNOT00000077672
|
LOC103692976
|
cyclin-T1-like |
| chr2_+_2729829 | 0.06 |
ENSRNOT00000017068
|
Spata9
|
spermatogenesis associated 9 |
| chr5_+_116421894 | 0.06 |
ENSRNOT00000080577
ENSRNOT00000086628 ENSRNOT00000004017 |
Nfia
|
nuclear factor I/A |
| chr2_+_196334626 | 0.06 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr6_-_125723732 | 0.06 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr6_-_51019407 | 0.06 |
ENSRNOT00000011659
|
Gpr22
|
G protein-coupled receptor 22 |
| chr6_-_67084234 | 0.06 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr1_-_275882444 | 0.06 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr6_-_8344574 | 0.05 |
ENSRNOT00000009660
|
Prepl
|
prolyl endopeptidase-like |
| chr15_+_11298478 | 0.05 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr11_-_1836897 | 0.05 |
ENSRNOT00000050533
|
Zfp654
|
zinc finger protein 654 |
| chr16_-_27472687 | 0.05 |
ENSRNOT00000046984
|
AABR07025074.1
|
|
| chr3_-_154490851 | 0.05 |
ENSRNOT00000017213
|
Tti1
|
TELO2 interacting protein 1 |
| chr3_-_103745236 | 0.05 |
ENSRNOT00000006876
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr13_-_98478327 | 0.05 |
ENSRNOT00000030135
|
Coq8a
|
coenzyme Q8A |
| chr10_+_97212432 | 0.05 |
ENSRNOT00000088599
|
Axin2
|
axin 2 |
| chr2_-_33025271 | 0.05 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr9_-_54351339 | 0.05 |
ENSRNOT00000080522
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr1_+_100299626 | 0.05 |
ENSRNOT00000092327
ENSRNOT00000044257 |
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr13_-_89242443 | 0.05 |
ENSRNOT00000029202
|
Atf6
|
activating transcription factor 6 |
| chr11_+_66713888 | 0.05 |
ENSRNOT00000003340
|
Fbxo40
|
F-box protein 40 |
| chr11_-_4397361 | 0.05 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr13_+_47454591 | 0.05 |
ENSRNOT00000005791
|
LOC498222
|
similar to specifically androgen-regulated protein |
| chr10_+_39850818 | 0.04 |
ENSRNOT00000012671
|
Fnip1
|
folliculin interacting protein 1 |
| chr16_+_60925093 | 0.04 |
ENSRNOT00000015813
|
Tnks
|
tankyrase |
| chr1_-_163611299 | 0.04 |
ENSRNOT00000043788
|
Emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr6_+_8219385 | 0.04 |
ENSRNOT00000040509
|
Ppm1b
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr4_-_183374195 | 0.04 |
ENSRNOT00000073927
ENSRNOT00000074740 ENSRNOT00000070907 |
Caprin2
|
caprin family member 2 |
| chr13_+_98311827 | 0.04 |
ENSRNOT00000082844
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr7_-_2941122 | 0.04 |
ENSRNOT00000082107
|
Esyt1
|
extended synaptotagmin 1 |
| chrX_-_23246719 | 0.04 |
ENSRNOT00000085588
|
AABR07037536.1
|
|
| chr18_+_30820321 | 0.04 |
ENSRNOT00000060472
|
Pcdhga3
|
protocadherin gamma subfamily A, 3 |
| chr11_-_62451149 | 0.04 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_+_214664537 | 0.04 |
ENSRNOT00000075002
|
AABR07006030.1
|
|
| chr12_+_22451596 | 0.04 |
ENSRNOT00000072838
|
Trip6
|
thyroid hormone receptor interactor 6 |
| chr8_-_46675544 | 0.04 |
ENSRNOT00000052293
|
Tecta
|
tectorin alpha |
| chr2_+_252771017 | 0.04 |
ENSRNOT00000075452
ENSRNOT00000055318 |
Ttll7
|
tubulin tyrosine ligase like 7 |
| chr16_+_8823872 | 0.03 |
ENSRNOT00000027186
|
Drgx
|
dorsal root ganglia homeobox |
| chrX_+_157759624 | 0.03 |
ENSRNOT00000003192
|
LOC686087
|
similar to motile sperm domain containing 1 |
| chr16_-_48437223 | 0.03 |
ENSRNOT00000013005
ENSRNOT00000059401 |
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr9_-_16581078 | 0.03 |
ENSRNOT00000022582
|
Pex6
|
peroxisomal biogenesis factor 6 |
| chr1_+_1784078 | 0.03 |
ENSRNOT00000020098
|
Lats1
|
large tumor suppressor kinase 1 |
| chr9_-_81566642 | 0.03 |
ENSRNOT00000080345
|
Aamp
|
angio-associated, migratory cell protein |
| chr1_+_154606490 | 0.03 |
ENSRNOT00000024095
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr12_+_49761120 | 0.03 |
ENSRNOT00000070961
|
Myo18b
|
myosin XVIIIb |
| chr12_-_5685448 | 0.03 |
ENSRNOT00000077167
|
Fry
|
FRY microtubule binding protein |
| chr3_-_71798531 | 0.03 |
ENSRNOT00000088170
|
Calcrl
|
calcitonin receptor like receptor |
| chr6_+_86823684 | 0.03 |
ENSRNOT00000086081
ENSRNOT00000006574 |
Fancm
|
Fanconi anemia, complementation group M |
| chr14_-_72025137 | 0.03 |
ENSRNOT00000080604
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr17_-_44643362 | 0.02 |
ENSRNOT00000091041
|
Zfp184
|
zinc finger protein 184 |
| chr18_+_30527705 | 0.02 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr10_+_55687050 | 0.02 |
ENSRNOT00000057136
|
Per1
|
period circadian clock 1 |
| chr8_-_130429132 | 0.02 |
ENSRNOT00000026261
|
Hhatl
|
hedgehog acyltransferase-like |
| chrX_+_76083549 | 0.02 |
ENSRNOT00000003573
|
Magee1
|
MAGE family member E1 |
| chr9_+_94316717 | 0.02 |
ENSRNOT00000077005
ENSRNOT00000092534 |
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr7_+_144080614 | 0.02 |
ENSRNOT00000077687
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr17_-_76250537 | 0.02 |
ENSRNOT00000092693
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr4_-_135069970 | 0.02 |
ENSRNOT00000008221
|
Cntn3
|
contactin 3 |
| chr2_+_149315189 | 0.02 |
ENSRNOT00000082464
|
Med12l
|
mediator complex subunit 12-like |
| chr2_-_177924970 | 0.02 |
ENSRNOT00000029340
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr5_+_148320438 | 0.02 |
ENSRNOT00000018742
|
Pef1
|
penta-EF hand domain containing 1 |
| chr18_+_64008048 | 0.02 |
ENSRNOT00000083414
|
Ldlrad4
|
low density lipoprotein receptor class A domain containing 4 |
| chrX_+_9436707 | 0.02 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr10_-_65502936 | 0.02 |
ENSRNOT00000089615
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr4_-_162498052 | 0.02 |
ENSRNOT00000066349
|
LOC100360260
|
chromobox homolog 3-like |
| chr4_+_34237762 | 0.02 |
ENSRNOT00000084163
|
Mios
|
meiosis regulator for oocyte development |
| chrX_+_70440059 | 0.02 |
ENSRNOT00000003946
|
Arr3
|
arrestin 3 |
| chr5_-_150704117 | 0.02 |
ENSRNOT00000067455
|
Sesn2
|
sestrin 2 |
| chr2_-_118882562 | 0.01 |
ENSRNOT00000058860
|
Kcnmb3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr1_+_224927764 | 0.01 |
ENSRNOT00000050814
|
AC099294.1
|
|
| chr2_+_68821004 | 0.01 |
ENSRNOT00000083713
|
Egf
|
epidermal growth factor |
| chr5_-_12199283 | 0.01 |
ENSRNOT00000007769
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr16_-_48692476 | 0.01 |
ENSRNOT00000013118
|
Irf2
|
interferon regulatory factor 2 |
| chr7_-_132984110 | 0.01 |
ENSRNOT00000046626
|
Hmgb1-ps3
|
high mobility group box 1, pseudogene 3 |
| chr4_+_183896303 | 0.01 |
ENSRNOT00000055437
|
RGD1309621
|
similar to hypothetical protein FLJ10652 |
| chr18_+_29386809 | 0.01 |
ENSRNOT00000082079
ENSRNOT00000024825 |
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr1_-_192057612 | 0.01 |
ENSRNOT00000024471
|
Ndufab1
|
NADH:ubiquinone oxidoreductase subunit AB1 |
| chr15_-_27819376 | 0.01 |
ENSRNOT00000067400
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr8_-_116438038 | 0.01 |
ENSRNOT00000023791
|
Gnat1
|
G protein subunit alpha transducin 1 |
| chr14_-_115391652 | 0.01 |
ENSRNOT00000002094
|
Chac2
|
ChaC cation transport regulator 2 |
| chr5_-_711033 | 0.01 |
ENSRNOT00000024199
|
Crispld1
|
cysteine-rich secretory protein LCCL domain containing 1 |
| chr20_+_3558827 | 0.01 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr8_+_103938520 | 0.01 |
ENSRNOT00000067835
|
Gk5
|
glycerol kinase 5 (putative) |
| chr2_+_38230757 | 0.01 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chr10_+_82032656 | 0.01 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr1_+_154446156 | 0.01 |
ENSRNOT00000093052
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr4_+_66091641 | 0.01 |
ENSRNOT00000043147
|
AABR07060287.1
|
|
| chr5_-_105582375 | 0.01 |
ENSRNOT00000083373
|
Slc24a2
|
solute carrier family 24 member 2 |
| chr6_+_80218880 | 0.01 |
ENSRNOT00000006995
|
Mia2
|
melanoma inhibitory activity 2 |
| chr5_+_25168295 | 0.01 |
ENSRNOT00000020245
|
Fsbp
|
fibrinogen silencer binding protein |
| chr6_+_73358112 | 0.01 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr20_-_6234703 | 0.01 |
ENSRNOT00000092453
|
Stk38
|
serine/threonine kinase 38 |
| chr14_-_45165207 | 0.01 |
ENSRNOT00000002960
|
Klf3
|
Kruppel like factor 3 |
| chr1_-_267463694 | 0.01 |
ENSRNOT00000084851
ENSRNOT00000016645 |
Col17a1
|
collagen type XVII alpha 1 chain |
| chr2_-_5577369 | 0.01 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr11_+_72044096 | 0.01 |
ENSRNOT00000034757
|
Senp5
|
SUMO1/sentrin specific peptidase 5 |
| chr8_+_94686938 | 0.01 |
ENSRNOT00000013285
|
Ripply2
|
ripply transcriptional repressor 2 |
| chr1_-_174620064 | 0.01 |
ENSRNOT00000016224
|
Tmem41b
|
transmembrane protein 41B |
| chrX_-_13279082 | 0.00 |
ENSRNOT00000051898
ENSRNOT00000060857 |
Tspan7
|
tetraspanin 7 |
| chr10_-_87248572 | 0.00 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr4_-_181348799 | 0.00 |
ENSRNOT00000033743
|
LOC690784
|
hypothetical protein LOC690783 |
| chr9_-_74048244 | 0.00 |
ENSRNOT00000018293
|
Lancl1
|
LanC like 1 |
| chr7_+_12619774 | 0.00 |
ENSRNOT00000015257
|
Med16
|
mediator complex subunit 16 |
| chr10_-_87529599 | 0.00 |
ENSRNOT00000074099
|
Krtap2-1
|
keratin associated protein 2-1 |
| chr1_+_225037737 | 0.00 |
ENSRNOT00000077959
|
Bscl2
|
BSCL2, seipin lipid droplet biogenesis associated |
| chr18_+_30509393 | 0.00 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chrX_-_42329232 | 0.00 |
ENSRNOT00000045705
|
LOC100361934
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 |
| chr2_-_31753528 | 0.00 |
ENSRNOT00000075057
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr10_+_78111050 | 0.00 |
ENSRNOT00000079364
|
Cox11
|
COX11 cytochrome c oxidase copper chaperone |
| chr4_+_129574264 | 0.00 |
ENSRNOT00000010185
|
Arl6ip5
|
ADP-ribosylation factor like GTPase 6 interacting protein 5 |
| chr3_-_63836017 | 0.00 |
ENSRNOT00000030978
|
AABR07052585.1
|
|
| chr19_+_56803382 | 0.00 |
ENSRNOT00000036042
|
Urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.2 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.1 | 0.2 | GO:0072641 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.1 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.0 | 0.3 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.1 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 0.0 | 0.0 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.0 | 0.1 | GO:0071245 | cellular response to carbon monoxide(GO:0071245) cellular response to nitrite(GO:0071250) response to nitrite(GO:0080033) |
| 0.0 | 0.1 | GO:0072268 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:1904742 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.0 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.0 | 0.0 | GO:0050893 | sensory processing(GO:0050893) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |