Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
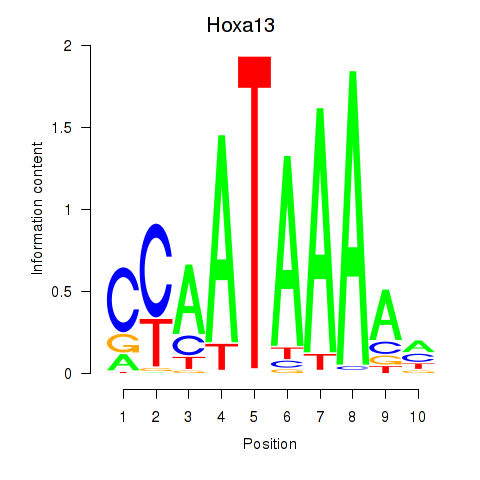
Results for Hoxa13
Z-value: 0.94
Transcription factors associated with Hoxa13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa13
|
ENSRNOG00000057061 | homeo box A13 |
Activity profile of Hoxa13 motif
Sorted Z-values of Hoxa13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_27862868 | 2.92 |
ENSRNOT00000004877
|
Gabra6
|
gamma-aminobutyric acid type A receptor alpha 6 subunit |
| chr4_+_170149029 | 0.85 |
ENSRNOT00000073287
|
LOC103690002
|
histone H2A.J |
| chr17_-_61332391 | 0.63 |
ENSRNOT00000034599
|
LOC100362965
|
SNRPN upstream reading frame protein-like |
| chr3_-_64554953 | 0.52 |
ENSRNOT00000067452
|
LOC102553814
|
serine/arginine repetitive matrix protein 1-like |
| chr20_-_5064469 | 0.52 |
ENSRNOT00000001120
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr2_+_198359754 | 0.44 |
ENSRNOT00000048582
|
Hist2h2ab
|
histone cluster 2 H2A family member b |
| chr10_-_59888198 | 0.43 |
ENSRNOT00000093482
ENSRNOT00000049311 ENSRNOT00000093230 |
Aspa
|
aspartoacylase |
| chr1_-_124803363 | 0.41 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chrX_+_33884499 | 0.36 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr7_-_136997050 | 0.36 |
ENSRNOT00000009143
|
Dbx2
|
developing brain homeobox 2 |
| chr4_-_727691 | 0.35 |
ENSRNOT00000008497
|
Shh
|
sonic hedgehog |
| chr3_+_79613171 | 0.34 |
ENSRNOT00000011467
|
Agbl2
|
ATP/GTP binding protein-like 2 |
| chr6_-_106971250 | 0.33 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr4_+_78263866 | 0.32 |
ENSRNOT00000033807
|
AI854703
|
expressed sequence AI854703 |
| chr10_-_40953467 | 0.31 |
ENSRNOT00000092189
|
Glra1
|
glycine receptor, alpha 1 |
| chr4_-_179512471 | 0.30 |
ENSRNOT00000012588
|
Kras
|
KRAS proto-oncogene, GTPase |
| chr19_-_37916813 | 0.29 |
ENSRNOT00000026585
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr2_-_198339130 | 0.28 |
ENSRNOT00000028767
|
Bola1
|
bolA family member 1 |
| chr2_-_210116038 | 0.28 |
ENSRNOT00000074950
|
LOC684509
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr7_+_123168811 | 0.27 |
ENSRNOT00000007091
|
Csdc2
|
cold shock domain containing C2 |
| chr2_-_198360678 | 0.27 |
ENSRNOT00000051917
|
Hist2h2ac
|
histone cluster 2 H2A family member c |
| chr5_+_139963002 | 0.27 |
ENSRNOT00000048506
|
Col9a2
|
collagen type IX alpha 2 chain |
| chr7_+_38742051 | 0.26 |
ENSRNOT00000006070
|
Dcn
|
decorin |
| chr16_+_54332660 | 0.26 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr17_-_78561613 | 0.25 |
ENSRNOT00000020220
|
Fam107b
|
family with sequence similarity 107, member B |
| chr13_+_36532758 | 0.25 |
ENSRNOT00000083007
|
En1
|
engrailed 1 |
| chr4_+_66276835 | 0.25 |
ENSRNOT00000007544
|
Fmc1
|
formation of mitochondrial complex V assembly factor 1 |
| chr4_+_157726941 | 0.25 |
ENSRNOT00000025081
|
Vamp1
|
vesicle-associated membrane protein 1 |
| chr19_+_25123724 | 0.24 |
ENSRNOT00000007407
|
LOC686013
|
hypothetical protein LOC686013 |
| chr15_-_23969011 | 0.23 |
ENSRNOT00000014821
|
Gch1
|
GTP cyclohydrolase 1 |
| chr4_-_10329241 | 0.23 |
ENSRNOT00000017232
|
Fgl2
|
fibrinogen-like 2 |
| chr18_+_70970596 | 0.21 |
ENSRNOT00000025217
ENSRNOT00000039203 |
Rpl17
|
ribosomal protein L17 |
| chr4_-_51199570 | 0.21 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr1_-_69835185 | 0.21 |
ENSRNOT00000020757
|
LOC108349745
|
spindle assembly abnormal protein 6 homolog |
| chr5_-_150167077 | 0.20 |
ENSRNOT00000088493
ENSRNOT00000013761 ENSRNOT00000067349 ENSRNOT00000051097 ENSRNOT00000067189 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr4_-_170860225 | 0.20 |
ENSRNOT00000007577
|
Mgp
|
matrix Gla protein |
| chr4_-_130659697 | 0.20 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr4_+_155313671 | 0.20 |
ENSRNOT00000020812
|
Mfap5
|
microfibrillar associated protein 5 |
| chr11_-_87081950 | 0.20 |
ENSRNOT00000002574
|
Dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr4_+_170766900 | 0.19 |
ENSRNOT00000044446
|
H2afj
|
H2A histone family, member J |
| chr9_-_79545024 | 0.19 |
ENSRNOT00000021152
|
Mreg
|
melanoregulin |
| chr10_-_56558487 | 0.19 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr3_+_131351587 | 0.19 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr3_+_79713567 | 0.19 |
ENSRNOT00000012110
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4 |
| chr13_-_61070599 | 0.18 |
ENSRNOT00000005251
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr11_-_46369577 | 0.18 |
ENSRNOT00000049859
ENSRNOT00000047817 ENSRNOT00000002220 ENSRNOT00000002223 ENSRNOT00000063864 ENSRNOT00000070871 ENSRNOT00000072140 |
Abi3bp
|
ABI family member 3 binding protein |
| chr1_-_93949187 | 0.17 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr9_+_98668231 | 0.17 |
ENSRNOT00000027528
|
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chrX_-_123474154 | 0.17 |
ENSRNOT00000092415
ENSRNOT00000092455 |
RGD1564541
|
similar to hypothetical protein FLJ22965 |
| chr10_+_54155876 | 0.17 |
ENSRNOT00000072855
|
Gas7
|
growth arrest specific 7 |
| chr8_+_22458899 | 0.17 |
ENSRNOT00000066007
ENSRNOT00000010947 ENSRNOT00000065028 |
Dnm2
|
dynamin 2 |
| chr11_+_72705129 | 0.17 |
ENSRNOT00000073330
|
Apod
|
apolipoprotein D |
| chr2_+_200603426 | 0.17 |
ENSRNOT00000072189
|
LOC102550892
|
zinc finger protein 697-like |
| chr4_-_82141385 | 0.17 |
ENSRNOT00000008447
|
Hoxa3
|
homeobox A3 |
| chr2_-_205212681 | 0.17 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr10_+_72272248 | 0.16 |
ENSRNOT00000003908
|
Car4
|
carbonic anhydrase 4 |
| chr1_+_124477164 | 0.16 |
ENSRNOT00000048744
|
Hmgn5b
|
high mobility group nucleosome binding domain 5B |
| chr11_+_80255790 | 0.16 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr2_+_200572502 | 0.16 |
ENSRNOT00000074666
|
Zfp697
|
zinc finger protein 697 |
| chr1_-_67065797 | 0.16 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chrX_-_14890606 | 0.16 |
ENSRNOT00000049864
|
RGD1560784
|
similar to RIKEN cDNA B630019K06 |
| chr2_+_38230757 | 0.16 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chr4_+_3959640 | 0.16 |
ENSRNOT00000009439
|
Paxip1
|
PAX interacting protein 1 |
| chr6_-_78817641 | 0.16 |
ENSRNOT00000072117
|
AABR07064392.1
|
|
| chr14_+_5928737 | 0.16 |
ENSRNOT00000071877
ENSRNOT00000040985 ENSRNOT00000074889 |
Mpa2l
|
macrophage activation 2 like |
| chr15_-_37831031 | 0.15 |
ENSRNOT00000091562
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr12_-_2438817 | 0.15 |
ENSRNOT00000037059
|
Ccl25
|
C-C motif chemokine ligand 25 |
| chr1_-_222242644 | 0.15 |
ENSRNOT00000050891
|
Vegfb
|
vascular endothelial growth factor B |
| chr17_+_9289189 | 0.15 |
ENSRNOT00000015632
|
H2afy
|
H2A histone family, member Y |
| chr2_-_208215187 | 0.15 |
ENSRNOT00000047836
|
Rap1a
|
RAP1A, member of RAS oncogene family |
| chr20_+_26534535 | 0.14 |
ENSRNOT00000000423
|
Tmem167b
|
transmembrane protein 167B |
| chr4_-_180234804 | 0.14 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr5_-_150949931 | 0.14 |
ENSRNOT00000067905
|
Smpdl3b
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr14_-_20920286 | 0.14 |
ENSRNOT00000004391
|
Slc4a4
|
solute carrier family 4 member 4 |
| chr4_+_180062799 | 0.14 |
ENSRNOT00000021623
|
Rassf8
|
Ras association domain family member 8 |
| chr7_-_74328603 | 0.14 |
ENSRNOT00000079159
|
Atmin
|
ATM interactor |
| chr15_-_76789298 | 0.14 |
ENSRNOT00000077853
|
AABR07018857.1
|
|
| chr17_-_80860195 | 0.14 |
ENSRNOT00000038651
|
Trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr2_+_8732932 | 0.14 |
ENSRNOT00000072849
|
Arrdc3
|
arrestin domain containing 3 |
| chr3_+_114772603 | 0.14 |
ENSRNOT00000073569
|
MGC105649
|
hypothetical LOC302884 |
| chr10_+_46314375 | 0.14 |
ENSRNOT00000088300
|
Med9
|
mediator complex subunit 9 |
| chr15_+_61673560 | 0.13 |
ENSRNOT00000093661
|
Mtrf1
|
mitochondrial translation release factor 1 |
| chr3_+_145764932 | 0.13 |
ENSRNOT00000075257
|
AABR07054264.1
|
|
| chrX_+_6273733 | 0.13 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr4_+_153217782 | 0.13 |
ENSRNOT00000015499
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr11_+_36634662 | 0.13 |
ENSRNOT00000050178
|
B3galt5
|
Beta-1,3-galactosyltransferase 5 |
| chr19_+_52515022 | 0.13 |
ENSRNOT00000022091
|
Klhl36
|
kelch-like family member 36 |
| chr9_-_9702306 | 0.13 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr20_+_248410 | 0.13 |
ENSRNOT00000049573
|
Tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr16_+_50152008 | 0.13 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr1_+_251633199 | 0.12 |
ENSRNOT00000084785
|
AC094647.2
|
|
| chr11_-_59110562 | 0.12 |
ENSRNOT00000047907
ENSRNOT00000042024 |
Lsamp
|
limbic system-associated membrane protein |
| chr1_-_89369960 | 0.12 |
ENSRNOT00000028545
|
Hamp
|
hepcidin antimicrobial peptide |
| chr20_+_1749716 | 0.12 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr16_-_20860767 | 0.12 |
ENSRNOT00000027275
ENSRNOT00000092417 |
Gdf1
Cers1
|
growth differentiation factor 1 ceramide synthase 1 |
| chr5_-_146973932 | 0.12 |
ENSRNOT00000007761
|
Zfp362
|
zinc finger protein 362 |
| chr3_-_8766433 | 0.12 |
ENSRNOT00000021865
|
Kyat1
|
kynurenine aminotransferase 1 |
| chrX_+_71528988 | 0.12 |
ENSRNOT00000075963
|
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr7_+_79638046 | 0.12 |
ENSRNOT00000029419
|
Zfpm2
|
zinc finger protein, multitype 2 |
| chr5_-_174921 | 0.12 |
ENSRNOT00000056239
|
Tipinl1
|
TIMELESS interacting protein like 1 |
| chr8_+_117906014 | 0.12 |
ENSRNOT00000056180
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr8_+_99568958 | 0.12 |
ENSRNOT00000073400
|
Plscr5
|
phospholipid scramblase family, member 5 |
| chr3_+_149624712 | 0.12 |
ENSRNOT00000018581
|
Bpifa1
|
BPI fold containing family A, member 1 |
| chr4_+_165732643 | 0.12 |
ENSRNOT00000034403
|
LOC690326
|
hypothetical protein LOC690326 |
| chr8_+_118066988 | 0.11 |
ENSRNOT00000056161
|
Map4
|
microtubule-associated protein 4 |
| chr5_-_113880911 | 0.11 |
ENSRNOT00000029441
|
Eqtn
|
equatorin |
| chr2_+_188583664 | 0.11 |
ENSRNOT00000045477
|
Dpm3
|
dolichyl-phosphate mannosyltransferase subunit 3 |
| chr3_+_61658245 | 0.11 |
ENSRNOT00000033511
|
Hoxd3
|
homeo box D3 |
| chr17_-_42127678 | 0.11 |
ENSRNOT00000024196
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr1_+_204959174 | 0.11 |
ENSRNOT00000023257
|
Zranb1
|
zinc finger RANBP2-type containing 1 |
| chr4_-_176679815 | 0.11 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr12_-_13668515 | 0.11 |
ENSRNOT00000086847
|
Fscn1
|
fascin actin-bundling protein 1 |
| chr3_+_160935709 | 0.11 |
ENSRNOT00000019688
|
Dbndd2
|
dysbindin domain containing 2 |
| chr1_+_279798187 | 0.11 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr20_+_6877091 | 0.11 |
ENSRNOT00000093654
|
RGD735065
|
similar to GI:13385412-like protein splice form I |
| chr5_-_139853247 | 0.11 |
ENSRNOT00000056644
|
Exo5
|
exonuclease 5 |
| chr18_+_3565166 | 0.11 |
ENSRNOT00000039377
|
Riok3
|
RIO kinase 3 |
| chrX_-_73766885 | 0.11 |
ENSRNOT00000091325
|
Rlim
|
ring finger protein, LIM domain interacting |
| chr10_+_58784660 | 0.11 |
ENSRNOT00000019925
|
LOC100912483
|
uncharacterized LOC100912483 |
| chr6_+_55812747 | 0.11 |
ENSRNOT00000008106
|
Sostdc1
|
sclerostin domain containing 1 |
| chr12_+_51878153 | 0.11 |
ENSRNOT00000056798
|
Hscb
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr2_-_96509424 | 0.10 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr7_-_70319346 | 0.10 |
ENSRNOT00000071543
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr6_+_56846789 | 0.10 |
ENSRNOT00000032108
|
Agmo
|
alkylglycerol monooxygenase |
| chr15_-_39705208 | 0.10 |
ENSRNOT00000015412
|
Phf11
|
PHD finger protein 11 |
| chr7_-_15856676 | 0.10 |
ENSRNOT00000086553
|
AABR07055919.1
|
|
| chr17_-_29895386 | 0.10 |
ENSRNOT00000048757
|
Cdyl
|
chromodomain Y-like |
| chr10_-_97756521 | 0.10 |
ENSRNOT00000045902
|
Slc16a6
|
solute carrier family 16, member 6 |
| chr6_+_137889261 | 0.10 |
ENSRNOT00000034995
|
Tex22
|
testis expressed 22 |
| chr1_+_207508414 | 0.10 |
ENSRNOT00000054896
|
Nps
|
neuropeptide S |
| chr13_+_90533365 | 0.10 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chrX_+_112769645 | 0.10 |
ENSRNOT00000064478
|
Col4a5
|
collagen type IV alpha 5 chain |
| chr2_+_196277978 | 0.10 |
ENSRNOT00000028625
|
Vps72
|
vacuolar protein sorting 72 homolog |
| chr5_+_144054452 | 0.10 |
ENSRNOT00000011565
|
Mrps15
|
mitochondrial ribosomal protein S15 |
| chr5_-_139748489 | 0.09 |
ENSRNOT00000078741
|
Nfyc
|
nuclear transcription factor Y subunit gamma |
| chr17_-_44643362 | 0.09 |
ENSRNOT00000091041
|
Zfp184
|
zinc finger protein 184 |
| chr3_-_64364101 | 0.09 |
ENSRNOT00000089065
|
Zfp385b
|
zinc finger protein 385B |
| chr1_+_197813963 | 0.09 |
ENSRNOT00000045531
|
LOC100361060
|
ribosomal protein L36-like |
| chr15_+_17775692 | 0.09 |
ENSRNOT00000061169
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr11_-_4397361 | 0.09 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr8_+_91820783 | 0.09 |
ENSRNOT00000043351
|
RGD1560917
|
similar to mitochondrial ribosomal protein L41 |
| chr8_-_96985558 | 0.09 |
ENSRNOT00000018553
|
LOC501033
|
similar to UPF0258 protein KIAA1024 |
| chr2_-_148807813 | 0.09 |
ENSRNOT00000047649
|
AC112531.1
|
|
| chr13_-_93677377 | 0.09 |
ENSRNOT00000004917
|
Fh
|
fumarate hydratase |
| chr4_-_6978073 | 0.09 |
ENSRNOT00000045311
|
Nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr10_-_47546345 | 0.09 |
ENSRNOT00000077461
|
Aldh3a2
|
aldehyde dehydrogenase 3 family, member A2 |
| chr20_+_2088533 | 0.09 |
ENSRNOT00000001012
ENSRNOT00000079021 |
Znrd1
|
zinc ribbon domain containing 1 |
| chr9_+_52023295 | 0.09 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr9_+_51298426 | 0.09 |
ENSRNOT00000065212
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr3_+_151126591 | 0.09 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr5_+_50381244 | 0.09 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chrX_+_90588124 | 0.08 |
ENSRNOT00000045685
|
Tgif2lx2
|
TGFB-induced factor homeobox 2-like, X-linked 2 |
| chrX_-_153080915 | 0.08 |
ENSRNOT00000091365
|
Xlr4a
|
X-linked lymphocyte-regulated 4A |
| chr3_+_108944141 | 0.08 |
ENSRNOT00000034950
|
Fam98b
|
family with sequence similarity 98, member B |
| chr4_+_64088900 | 0.08 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr10_-_62184874 | 0.08 |
ENSRNOT00000004229
|
Rpa1
|
replication protein A1 |
| chr1_-_89483988 | 0.08 |
ENSRNOT00000028603
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr18_+_79406381 | 0.08 |
ENSRNOT00000022303
ENSRNOT00000058295 ENSRNOT00000058296 ENSRNOT00000022280 |
Mbp
|
myelin basic protein |
| chr18_+_29951094 | 0.08 |
ENSRNOT00000027402
|
LOC102553180
|
protocadherin alpha-1-like |
| chr5_+_5279296 | 0.08 |
ENSRNOT00000045373
|
Tram1
|
translocation associated membrane protein 1 |
| chr4_-_157486844 | 0.08 |
ENSRNOT00000038281
ENSRNOT00000022874 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr7_+_35309265 | 0.08 |
ENSRNOT00000080291
|
Tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr6_+_72124417 | 0.08 |
ENSRNOT00000040548
|
Scfd1
|
sec1 family domain containing 1 |
| chr17_-_23792353 | 0.08 |
ENSRNOT00000019089
|
Tbc1d7
|
TBC1 domain family, member 7 |
| chr10_+_80953006 | 0.08 |
ENSRNOT00000035567
|
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr2_+_237755673 | 0.08 |
ENSRNOT00000083937
|
Tbck
|
TBC1 domain containing kinase |
| chr11_+_7422272 | 0.08 |
ENSRNOT00000075964
|
Gbe1
|
1,4-alpha-glucan branching enzyme 1 |
| chr9_+_94324793 | 0.08 |
ENSRNOT00000092493
|
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr11_+_68198709 | 0.08 |
ENSRNOT00000003048
|
Dirc2
|
disrupted in renal carcinoma 2 |
| chr2_-_88553086 | 0.08 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr9_+_73529612 | 0.07 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr4_+_61850348 | 0.07 |
ENSRNOT00000013423
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr19_-_43911057 | 0.07 |
ENSRNOT00000026017
|
Ctrb1
|
chymotrypsinogen B1 |
| chr13_+_53283855 | 0.07 |
ENSRNOT00000043590
|
RGD1564839
|
similar to ribosomal protein L31 |
| chr2_+_30685840 | 0.07 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chr2_+_68821004 | 0.07 |
ENSRNOT00000083713
|
Egf
|
epidermal growth factor |
| chr3_-_40477754 | 0.07 |
ENSRNOT00000091168
|
AABR07052166.1
|
|
| chr7_-_73883812 | 0.07 |
ENSRNOT00000065722
|
Stk3
|
serine/threonine kinase 3 |
| chr3_-_154490851 | 0.07 |
ENSRNOT00000017213
|
Tti1
|
TELO2 interacting protein 1 |
| chr9_-_18371856 | 0.07 |
ENSRNOT00000027293
|
Supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chrX_-_1718637 | 0.07 |
ENSRNOT00000064309
|
Cdk16
|
cyclin-dependent kinase 16 |
| chr11_+_85508300 | 0.07 |
ENSRNOT00000038646
|
AABR07034730.3
|
|
| chr11_+_46184871 | 0.07 |
ENSRNOT00000048417
|
Tfg
|
Trk-fused gene |
| chr18_+_35489274 | 0.07 |
ENSRNOT00000073952
ENSRNOT00000076060 |
Dcp2
|
decapping mRNA 2 |
| chr3_-_151224123 | 0.07 |
ENSRNOT00000026091
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr7_-_11266630 | 0.07 |
ENSRNOT00000027926
|
Cactin
|
cactin, spliceosome C complex subunit |
| chr1_+_218003018 | 0.07 |
ENSRNOT00000028343
|
Fgf3
|
fibroblast growth factor 3 |
| chr10_+_764421 | 0.07 |
ENSRNOT00000084608
ENSRNOT00000087567 |
Myh11
|
myosin heavy chain 11 |
| chr9_-_88534710 | 0.07 |
ENSRNOT00000020880
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr4_+_162493908 | 0.07 |
ENSRNOT00000072064
|
Clec2d
|
C-type lectin domain family 2, member D |
| chr14_+_82769642 | 0.07 |
ENSRNOT00000065393
|
Ctbp1
|
C-terminal binding protein 1 |
| chr2_-_45518502 | 0.07 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr1_-_266862842 | 0.07 |
ENSRNOT00000027496
|
LOC103693430
|
up-regulated during skeletal muscle growth protein 5 |
| chr1_-_227932603 | 0.07 |
ENSRNOT00000033795
|
Ms4a6a
|
membrane spanning 4-domains A6A |
| chr2_+_23770721 | 0.07 |
ENSRNOT00000014795
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr15_+_48030973 | 0.07 |
ENSRNOT00000085487
ENSRNOT00000029757 |
Kif13b
|
kinesin family member 13B |
| chr16_-_20873344 | 0.07 |
ENSRNOT00000027381
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr14_+_84211600 | 0.07 |
ENSRNOT00000005996
|
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr19_+_38643108 | 0.06 |
ENSRNOT00000087392
|
LOC100360619
|
ribosomal protein L28-like |
| chr7_-_11406771 | 0.06 |
ENSRNOT00000047450
|
Eef2
|
eukaryotic translation elongation factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0065001 | positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) specification of axis polarity(GO:0065001) negative regulation of cholesterol efflux(GO:0090370) negative regulation of dopaminergic neuron differentiation(GO:1904339) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.1 | GO:0071282 | cellular response to iron(II) ion(GO:0071282) |
| 0.1 | 0.3 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.2 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.1 | 2.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.3 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.2 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 0.0 | 0.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:1900095 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.3 | GO:0034370 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) high-density lipoprotein particle assembly(GO:0034380) |
| 0.0 | 0.1 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.0 | 0.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.6 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.0 | GO:0021897 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.0 | 1.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of retinal cell programmed cell death(GO:0046671) cellular response to fluoride(GO:1902618) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.0 | GO:0042197 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.0 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.0 | GO:1903422 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.0 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.2 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.1 | GO:0044332 | STAT protein import into nucleus(GO:0007262) Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.2 | GO:0070821 | azurophil granule membrane(GO:0035577) tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.4 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.1 | 0.3 | GO:0030977 | taurine binding(GO:0030977) |
| 0.1 | 0.3 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 0.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.0 | GO:0031997 | N-terminal myristoylation domain binding(GO:0031997) calcium ion binding involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099567) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |