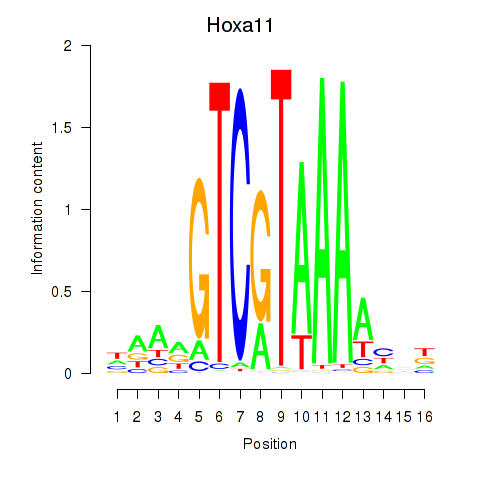
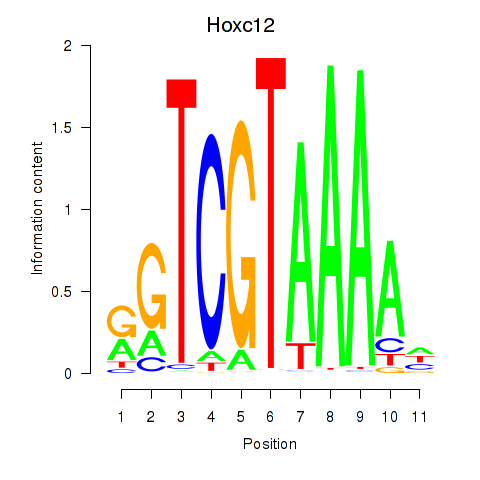
Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Hoxa11_Hoxc12
Z-value: 0.38
Transcription factors associated with Hoxa11_Hoxc12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa11
|
ENSRNOG00000059870 | homeobox A11 |
|
Hoxa11
|
ENSRNOG00000053884 | homeobox A11 |
|
Hoxc12
|
ENSRNOG00000016116 | homeo box C12 |
Activity profile of Hoxa11_Hoxc12 motif
Sorted Z-values of Hoxa11_Hoxc12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_4362717 | 0.28 |
ENSRNOT00000002887
|
Barhl2
|
BarH-like homeobox 2 |
| chr4_-_82173207 | 0.22 |
ENSRNOT00000074167
|
Hoxa5
|
homeo box A5 |
| chr7_-_13751271 | 0.20 |
ENSRNOT00000009931
|
Slc1a6
|
solute carrier family 1 member 6 |
| chr10_+_84152152 | 0.19 |
ENSRNOT00000010724
|
Hoxb5
|
homeo box B5 |
| chr5_+_152533349 | 0.13 |
ENSRNOT00000067524
|
Trim63
|
tripartite motif containing 63 |
| chr5_-_156734541 | 0.10 |
ENSRNOT00000021036
|
LOC100909857
|
cytidine deaminase-like |
| chr3_-_64554953 | 0.10 |
ENSRNOT00000067452
|
LOC102553814
|
serine/arginine repetitive matrix protein 1-like |
| chr15_-_37831031 | 0.09 |
ENSRNOT00000091562
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr1_+_154606490 | 0.08 |
ENSRNOT00000024095
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr4_+_66276835 | 0.08 |
ENSRNOT00000007544
|
Fmc1
|
formation of mitochondrial complex V assembly factor 1 |
| chr10_-_109622745 | 0.08 |
ENSRNOT00000074354
ENSRNOT00000088317 |
Pde6g
|
phosphodiesterase 6G |
| chr3_+_11593655 | 0.07 |
ENSRNOT00000074122
|
Pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr1_-_93949187 | 0.07 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr7_-_11963268 | 0.07 |
ENSRNOT00000080342
ENSRNOT00000025266 |
Csnk1g2
|
casein kinase 1, gamma 2 |
| chr4_-_34194764 | 0.07 |
ENSRNOT00000045270
|
Col28a1
|
collagen type XXVIII alpha 1 chain |
| chr13_+_47440803 | 0.07 |
ENSRNOT00000030476
|
Yod1
|
YOD1 deubiquitinase |
| chr4_+_10748745 | 0.07 |
ENSRNOT00000046142
|
Rsbn1l
|
round spermatid basic protein 1-like |
| chr17_+_44763598 | 0.06 |
ENSRNOT00000079880
|
Hist1h3b
|
histone cluster 1, H3b |
| chr6_-_15191660 | 0.06 |
ENSRNOT00000092654
|
Nrxn1
|
neurexin 1 |
| chr14_-_20920286 | 0.06 |
ENSRNOT00000004391
|
Slc4a4
|
solute carrier family 4 member 4 |
| chr5_-_147584038 | 0.06 |
ENSRNOT00000010983
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chr1_-_156327352 | 0.06 |
ENSRNOT00000074282
|
LOC100911776
|
coiled-coil domain-containing protein 89-like |
| chr4_+_97657671 | 0.06 |
ENSRNOT00000075400
|
Gng12
|
G protein subunit gamma 12 |
| chr8_+_82380757 | 0.06 |
ENSRNOT00000013512
|
Leo1
|
LEO1 homolog, Paf1/RNA polymerase II complex component |
| chr4_+_140247313 | 0.06 |
ENSRNOT00000040255
ENSRNOT00000064025 ENSRNOT00000041130 ENSRNOT00000043646 |
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr4_-_82258765 | 0.05 |
ENSRNOT00000008523
|
Hoxa5
|
homeo box A5 |
| chr17_+_77195247 | 0.05 |
ENSRNOT00000081940
|
Optn
|
optineurin |
| chr8_+_58347736 | 0.05 |
ENSRNOT00000080227
ENSRNOT00000066222 |
Slc35f2
|
solute carrier family 35, member F2 |
| chr1_-_53014466 | 0.05 |
ENSRNOT00000049831
|
Sft2d1
|
SFT2 domain containing 1 |
| chr9_+_50526811 | 0.05 |
ENSRNOT00000036990
|
RGD1305645
|
similar to RIKEN cDNA 1500015O10 |
| chr8_+_67615635 | 0.05 |
ENSRNOT00000008971
|
Itga11
|
integrin subunit alpha 11 |
| chr11_-_74866451 | 0.04 |
ENSRNOT00000002330
|
Il13ra1
|
interleukin 13 receptor subunit alpha 1 |
| chr2_+_159138758 | 0.04 |
ENSRNOT00000087942
|
AABR07011152.1
|
|
| chr11_-_4397361 | 0.04 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr17_+_9289189 | 0.04 |
ENSRNOT00000015632
|
H2afy
|
H2A histone family, member Y |
| chr1_+_99561523 | 0.04 |
ENSRNOT00000038229
|
Ceacam18
|
carcinoembryonic antigen-related cell adhesion molecule 18 |
| chr18_+_38847632 | 0.04 |
ENSRNOT00000014916
|
LOC102554034
|
ELMO domain-containing protein 2-like |
| chrX_+_111735820 | 0.04 |
ENSRNOT00000086948
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr14_+_84211600 | 0.04 |
ENSRNOT00000005996
|
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr7_+_121480723 | 0.04 |
ENSRNOT00000065304
|
Atf4
|
activating transcription factor 4 |
| chr7_-_119200623 | 0.04 |
ENSRNOT00000029753
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr8_-_55087832 | 0.04 |
ENSRNOT00000032152
|
Dlat
|
dihydrolipoamide S-acetyltransferase |
| chr14_-_3288017 | 0.03 |
ENSRNOT00000080452
|
LOC689986
|
hypothetical protein LOC689986 |
| chr14_+_46001849 | 0.03 |
ENSRNOT00000076611
|
Rell1
|
RELT-like 1 |
| chr1_+_128606770 | 0.03 |
ENSRNOT00000073355
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr10_-_13446135 | 0.03 |
ENSRNOT00000084991
|
Kctd5
|
potassium channel tetramerization domain containing 5 |
| chr7_+_117434419 | 0.03 |
ENSRNOT00000040981
|
Hgh1
|
HGH1 homolog |
| chr18_+_70263359 | 0.03 |
ENSRNOT00000019573
|
Cfap53
|
cilia and flagella associated protein 53 |
| chr6_-_51018050 | 0.03 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr8_+_99568958 | 0.03 |
ENSRNOT00000073400
|
Plscr5
|
phospholipid scramblase family, member 5 |
| chr18_+_36713869 | 0.03 |
ENSRNOT00000025465
|
Pou4f3
|
POU class 4 homeobox 3 |
| chr1_-_211265161 | 0.03 |
ENSRNOT00000080041
ENSRNOT00000023477 |
Bnip3
|
BCL2 interacting protein 3 |
| chr2_-_32518643 | 0.03 |
ENSRNOT00000061032
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chrX_+_131617798 | 0.03 |
ENSRNOT00000074384
|
Prr32
|
proline rich 32 |
| chr15_+_33544312 | 0.02 |
ENSRNOT00000020409
|
Bcl2l2
|
Bcl2-like 2 |
| chr6_+_96834525 | 0.02 |
ENSRNOT00000077935
|
Hif1a
|
hypoxia inducible factor 1 alpha subunit |
| chr1_-_225952516 | 0.02 |
ENSRNOT00000043387
|
Incenp
|
inner centromere protein |
| chr3_-_154490851 | 0.02 |
ENSRNOT00000017213
|
Tti1
|
TELO2 interacting protein 1 |
| chr2_-_18565842 | 0.02 |
ENSRNOT00000067456
ENSRNOT00000063821 ENSRNOT00000045532 |
Vcan
|
versican |
| chrX_-_2116483 | 0.02 |
ENSRNOT00000055077
|
Rp2
|
retinitis pigmentosa 2 (X-linked recessive) |
| chr3_-_152179193 | 0.02 |
ENSRNOT00000026700
|
Rbm12
|
RNA binding motif protein 12 |
| chr10_-_84920886 | 0.02 |
ENSRNOT00000068083
|
Sp2
|
Sp2 transcription factor |
| chr4_-_176679815 | 0.02 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr6_+_107531528 | 0.02 |
ENSRNOT00000077555
|
Acot3
|
acyl-CoA thioesterase 3 |
| chr14_-_44375804 | 0.02 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr5_-_60559533 | 0.02 |
ENSRNOT00000092899
|
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr3_-_121226125 | 0.02 |
ENSRNOT00000055875
|
Anapc1
|
anaphase promoting complex subunit 1 |
| chr1_-_53038229 | 0.02 |
ENSRNOT00000017282
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr5_+_165415136 | 0.02 |
ENSRNOT00000016317
ENSRNOT00000079407 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr5_-_109651730 | 0.02 |
ENSRNOT00000093032
|
Elavl2
|
ELAV like RNA binding protein 2 |
| chr2_-_202816562 | 0.02 |
ENSRNOT00000020401
|
Fam46c
|
family with sequence similarity 46, member C |
| chr3_+_159392193 | 0.02 |
ENSRNOT00000078101
ENSRNOT00000066260 |
Ift52
|
intraflagellar transport 52 |
| chr7_-_15852930 | 0.02 |
ENSRNOT00000009270
|
LOC691422
|
similar to zinc finger protein 101 |
| chr5_-_156781291 | 0.02 |
ENSRNOT00000075128
|
Cda
|
cytidine deaminase |
| chr9_-_17880706 | 0.02 |
ENSRNOT00000031549
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr7_-_91538673 | 0.02 |
ENSRNOT00000006209
|
Rad21
|
RAD21 cohesin complex component |
| chr16_+_62251243 | 0.02 |
ENSRNOT00000020347
|
Ubxn8
|
UBX domain protein 8 |
| chr16_-_20511818 | 0.02 |
ENSRNOT00000026567
|
Lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_230901126 | 0.01 |
ENSRNOT00000016026
ENSRNOT00000015564 ENSRNOT00000068198 |
Camk2d
|
calcium/calmodulin-dependent protein kinase II delta |
| chr15_-_39705208 | 0.01 |
ENSRNOT00000015412
|
Phf11
|
PHD finger protein 11 |
| chr4_-_50312608 | 0.01 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr8_+_99625545 | 0.01 |
ENSRNOT00000010689
ENSRNOT00000056727 |
Plscr1
|
phospholipid scramblase 1 |
| chrX_-_2116656 | 0.01 |
ENSRNOT00000081913
|
Rp2
|
retinitis pigmentosa 2 (X-linked recessive) |
| chr14_-_6201002 | 0.01 |
ENSRNOT00000049165
|
LOC100366030
|
rCG37858-like |
| chr8_-_36388224 | 0.01 |
ENSRNOT00000038312
|
Tirap
|
TIR domain containing adaptor protein |
| chr7_+_119554354 | 0.01 |
ENSRNOT00000000203
|
Csf2rb
|
colony stimulating factor 2 receptor beta common subunit |
| chr1_+_47162394 | 0.01 |
ENSRNOT00000068457
|
Tmem181
|
transmembrane protein 181 |
| chr16_-_64745207 | 0.01 |
ENSRNOT00000032467
|
Tti2
|
TELO2 interacting protein 2 |
| chr1_-_22625204 | 0.01 |
ENSRNOT00000021694
|
Vnn1
|
vanin 1 |
| chr11_-_70211701 | 0.01 |
ENSRNOT00000076114
|
Muc13
|
mucin 13, cell surface associated |
| chr2_-_30634243 | 0.01 |
ENSRNOT00000077537
|
Marveld2
|
MARVEL domain containing 2 |
| chr9_+_11031841 | 0.01 |
ENSRNOT00000074695
|
Sh3gl1
|
SH3 domain-containing GRB2-like 1 |
| chr5_-_14356692 | 0.01 |
ENSRNOT00000085654
|
Atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr2_+_180936688 | 0.01 |
ENSRNOT00000016039
|
Asic5
|
acid sensing ion channel subunit family member 5 |
| chr3_-_79282493 | 0.01 |
ENSRNOT00000049832
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr10_+_57131386 | 0.01 |
ENSRNOT00000026507
|
Psmb6
|
proteasome subunit beta 6 |
| chr7_+_118502676 | 0.01 |
ENSRNOT00000005925
|
Commd5
|
COMM domain containing 5 |
| chr7_+_54756043 | 0.01 |
ENSRNOT00000063917
|
Krr1
|
KRR1, small subunit processome component homolog |
| chr11_+_66316606 | 0.01 |
ENSRNOT00000041715
|
Stxbp5l
|
syntaxin binding protein 5-like |
| chr6_-_102372611 | 0.01 |
ENSRNOT00000085927
|
Rdh11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr7_+_66717434 | 0.01 |
ENSRNOT00000077878
|
Mon2
|
MON2 homolog, regulator of endosome-to-Golgi trafficking |
| chr18_-_37776453 | 0.01 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr16_+_80904125 | 0.01 |
ENSRNOT00000080832
|
Tmco3
|
transmembrane and coiled-coil domains 3 |
| chr20_-_30749940 | 0.01 |
ENSRNOT00000065127
|
Sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr2_+_196655469 | 0.01 |
ENSRNOT00000028730
|
Ctsk
|
cathepsin K |
| chr8_+_1459526 | 0.01 |
ENSRNOT00000034503
|
Kbtbd3
|
kelch repeat and BTB domain containing 3 |
| chr1_+_68239314 | 0.01 |
ENSRNOT00000070823
|
Vom1r109
|
vomeronasal 1 receptor 109 |
| chr5_+_127489418 | 0.00 |
ENSRNOT00000065275
|
RGD1559786
|
similar to RIKEN cDNA 0610037L13 |
| chr1_+_94718402 | 0.00 |
ENSRNOT00000046035
|
Uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr10_-_83128297 | 0.00 |
ENSRNOT00000082160
|
Kat7
|
lysine acetyltransferase 7 |
| chr1_-_271275989 | 0.00 |
ENSRNOT00000075570
|
LOC100912218
|
inositol 1,4,5-trisphosphate receptor-interacting protein-like |
| chr16_+_68633720 | 0.00 |
ENSRNOT00000081838
|
LOC100911229
|
sperm motility kinase-like |
| chr6_-_102372385 | 0.00 |
ENSRNOT00000078436
|
Rdh11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr10_-_90356242 | 0.00 |
ENSRNOT00000028496
|
Slc25a39
|
solute carrier family 25, member 39 |
| chr13_-_53870428 | 0.00 |
ENSRNOT00000000812
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr5_+_122019301 | 0.00 |
ENSRNOT00000068158
|
Pde4b
|
phosphodiesterase 4B |
| chr16_-_36080191 | 0.00 |
ENSRNOT00000017635
|
Hmgb2l1
|
high mobility group box 2-like 1 |
| chr13_-_47440682 | 0.00 |
ENSRNOT00000037679
ENSRNOT00000005729 ENSRNOT00000050354 ENSRNOT00000050859 |
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr6_-_92007917 | 0.00 |
ENSRNOT00000006425
ENSRNOT00000089891 |
Sos2
|
SOS Ras/Rho guanine nucleotide exchange factor 2 |
| chr11_+_73693814 | 0.00 |
ENSRNOT00000081081
ENSRNOT00000002354 ENSRNOT00000090940 |
Lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr10_+_63884338 | 0.00 |
ENSRNOT00000007100
|
Ywhae
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon |
| chr9_-_112027155 | 0.00 |
ENSRNOT00000021258
ENSRNOT00000080962 |
Pja2
|
praja ring finger ubiquitin ligase 2 |
| chr20_-_6556350 | 0.00 |
ENSRNOT00000035819
|
Lemd2
|
LEM domain containing 2 |
| chr13_-_51297621 | 0.00 |
ENSRNOT00000030926
|
Ndufv3
|
NADH:ubiquinone oxidoreductase subunit V3 |
| chr5_-_160282810 | 0.00 |
ENSRNOT00000076160
ENSRNOT00000016352 |
Ddi2
|
DNA-damage inducible protein 2 |
| chr6_-_51019407 | 0.00 |
ENSRNOT00000011659
|
Gpr22
|
G protein-coupled receptor 22 |
| chr2_-_189333322 | 0.00 |
ENSRNOT00000073599
ENSRNOT00000071253 ENSRNOT00000076325 |
Hax1
|
HCLS1 associated protein X-1 |
| chr17_+_47302272 | 0.00 |
ENSRNOT00000090021
|
Nme8
|
NME/NM23 family member 8 |
| chrX_-_153491837 | 0.00 |
ENSRNOT00000089027
|
LOC100360413
|
eukaryotic translation elongation factor 1 alpha 1-like |
| chr16_+_24980723 | 0.00 |
ENSRNOT00000082142
|
Tma16
|
translation machinery associated 16 homolog |
| chr5_-_115387377 | 0.00 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr1_+_247228061 | 0.00 |
ENSRNOT00000020809
|
Rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chr1_-_174379566 | 0.00 |
ENSRNOT00000067388
|
Tmem9b
|
TMEM9 domain family, member B |
| chr2_+_166856784 | 0.00 |
ENSRNOT00000035765
|
Otol1
|
otolin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa11_Hoxc12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.0 | GO:1990737 | negative regulation of translational initiation in response to stress(GO:0032057) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.0 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.0 | GO:0071250 | cellular response to nitrite(GO:0071250) response to nitrite(GO:0080033) |
| 0.0 | 0.0 | GO:0035772 | interleukin-13-mediated signaling pathway(GO:0035772) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:1990590 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |