Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Hoxa10
Z-value: 0.32
Transcription factors associated with Hoxa10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
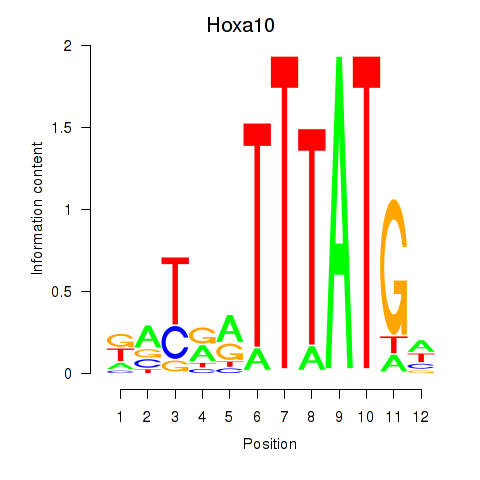
Hoxa10
|
ENSRNOG00000057650 | homeobox A10 |
Activity profile of Hoxa10 motif
Sorted Z-values of Hoxa10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_64224861 | 0.16 |
ENSRNOT00000093159
ENSRNOT00000093664 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr10_+_14136883 | 0.15 |
ENSRNOT00000082295
|
AC115181.1
|
|
| chr13_-_83425641 | 0.14 |
ENSRNOT00000063870
|
Tbx19
|
T-box 19 |
| chr2_+_231941794 | 0.11 |
ENSRNOT00000091475
|
AABR07013288.4
|
|
| chr8_+_130366775 | 0.11 |
ENSRNOT00000071152
|
Nktr
|
natural killer cell triggering receptor |
| chr6_+_106052212 | 0.10 |
ENSRNOT00000010626
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr9_-_65736852 | 0.10 |
ENSRNOT00000087458
|
Trak2
|
trafficking kinesin protein 2 |
| chr18_-_32670665 | 0.10 |
ENSRNOT00000019409
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr7_-_101138860 | 0.10 |
ENSRNOT00000077137
|
AABR07058124.4
|
|
| chr8_-_94920441 | 0.10 |
ENSRNOT00000014165
|
Cep162
|
centrosomal protein 162 |
| chr3_-_119405453 | 0.09 |
ENSRNOT00000090355
|
Sppl2a
|
signal peptide peptidase-like 2A |
| chr3_-_146396299 | 0.09 |
ENSRNOT00000040188
ENSRNOT00000008931 |
Apmap
|
adipocyte plasma membrane associated protein |
| chr18_+_30904498 | 0.09 |
ENSRNOT00000026969
|
Pcdhga11
|
protocadherin gamma subfamily A, 11 |
| chr10_-_27179254 | 0.09 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr4_-_125958112 | 0.09 |
ENSRNOT00000071315
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr9_-_65737538 | 0.09 |
ENSRNOT00000014939
|
Trak2
|
trafficking kinesin protein 2 |
| chr1_-_246010594 | 0.09 |
ENSRNOT00000023762
|
Rfx3
|
regulatory factor X3 |
| chr2_+_242882306 | 0.08 |
ENSRNOT00000013661
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr7_+_133457255 | 0.08 |
ENSRNOT00000080621
|
Cntn1
|
contactin 1 |
| chr15_-_76789298 | 0.08 |
ENSRNOT00000077853
|
AABR07018857.1
|
|
| chr2_-_40386669 | 0.08 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr14_-_82171480 | 0.07 |
ENSRNOT00000021952
|
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr16_-_9658484 | 0.07 |
ENSRNOT00000065216
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr14_-_28856214 | 0.07 |
ENSRNOT00000044659
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr13_+_98231326 | 0.07 |
ENSRNOT00000003837
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr3_+_113918629 | 0.06 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr3_-_66279155 | 0.06 |
ENSRNOT00000079887
|
Cerkl
|
ceramide kinase-like |
| chr1_-_90151405 | 0.06 |
ENSRNOT00000028688
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr12_+_38368693 | 0.06 |
ENSRNOT00000083261
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr19_-_22194740 | 0.06 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr17_-_86657473 | 0.06 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
| chr9_-_43454078 | 0.06 |
ENSRNOT00000023550
|
Tmem131
|
transmembrane protein 131 |
| chr10_-_104482838 | 0.05 |
ENSRNOT00000007246
|
Recql5
|
RecQ like helicase 5 |
| chr18_+_30496318 | 0.05 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr18_-_77317969 | 0.05 |
ENSRNOT00000090369
|
Nfatc1
|
nuclear factor of activated T-cells 1 |
| chr2_+_123597340 | 0.05 |
ENSRNOT00000058552
|
RGD1307100
|
similar to RIKEN cDNA D630029K19 |
| chr3_+_112390106 | 0.05 |
ENSRNOT00000081197
|
LOC691918
|
similar to Centrosomal protein of 27 kDa (Cep27 protein) |
| chr16_+_54319377 | 0.05 |
ENSRNOT00000090266
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr13_-_51784639 | 0.05 |
ENSRNOT00000089068
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr7_-_104801045 | 0.05 |
ENSRNOT00000079524
|
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr8_+_122197027 | 0.05 |
ENSRNOT00000013050
|
Ubp1
|
upstream binding protein 1 (LBP-1a) |
| chr17_+_81922329 | 0.04 |
ENSRNOT00000031542
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr2_+_4989295 | 0.04 |
ENSRNOT00000041541
|
Fam172a
|
family with sequence similarity 172, member A |
| chr9_-_52238564 | 0.04 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr5_+_116420690 | 0.04 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr11_+_62584959 | 0.04 |
ENSRNOT00000071065
|
Gramd1c
|
GRAM domain containing 1C |
| chr14_-_21128505 | 0.04 |
ENSRNOT00000004776
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr3_-_119405300 | 0.04 |
ENSRNOT00000015568
|
Sppl2a
|
signal peptide peptidase-like 2A |
| chr15_-_13228607 | 0.04 |
ENSRNOT00000042010
ENSRNOT00000088214 |
Ptprg
|
protein tyrosine phosphatase, receptor type, G |
| chr14_+_39964588 | 0.04 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr3_-_71798531 | 0.04 |
ENSRNOT00000088170
|
Calcrl
|
calcitonin receptor like receptor |
| chr1_-_250626844 | 0.04 |
ENSRNOT00000077135
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr2_+_231884337 | 0.04 |
ENSRNOT00000014695
|
Zgrf1
|
zinc finger, GRF-type containing 1 |
| chr3_-_25212049 | 0.04 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr5_+_147257678 | 0.04 |
ENSRNOT00000066268
|
Rnf19b
|
ring finger protein 19B |
| chr3_-_176197753 | 0.03 |
ENSRNOT00000013189
|
Dido1
|
death inducer-obliterator 1 |
| chr4_+_160020472 | 0.03 |
ENSRNOT00000078802
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr8_-_58253688 | 0.03 |
ENSRNOT00000010956
|
Cul5
|
cullin 5 |
| chr17_+_26478315 | 0.03 |
ENSRNOT00000021669
|
Slc35b3
|
solute carrier family 35 member B3 |
| chr7_+_74350479 | 0.03 |
ENSRNOT00000089034
|
AABR07057495.1
|
|
| chr5_-_58198782 | 0.03 |
ENSRNOT00000023951
|
Ccl21
|
C-C motif chemokine ligand 21 |
| chr7_-_122329443 | 0.03 |
ENSRNOT00000032232
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr16_-_2602103 | 0.03 |
ENSRNOT00000018413
|
Appl1
|
adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 |
| chr11_-_37253776 | 0.03 |
ENSRNOT00000002215
|
Dscam
|
DS cell adhesion molecule |
| chr7_+_132378273 | 0.03 |
ENSRNOT00000010990
|
LOC690142
|
hypothetical protein LOC690142 |
| chr13_-_82295123 | 0.03 |
ENSRNOT00000090120
|
LOC498265
|
similar to hypothetical protein FLJ10706 |
| chr2_-_147819335 | 0.03 |
ENSRNOT00000057909
|
Ankub1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr5_-_128839433 | 0.03 |
ENSRNOT00000065280
|
Osbpl9
|
oxysterol binding protein-like 9 |
| chr17_-_18421861 | 0.03 |
ENSRNOT00000060500
ENSRNOT00000036560 |
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr19_+_56803382 | 0.03 |
ENSRNOT00000036042
|
Urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr15_+_39644851 | 0.03 |
ENSRNOT00000081373
|
Rcbtb1
|
RCC1 and BTB domain containing protein 1 |
| chr14_+_42007312 | 0.03 |
ENSRNOT00000063985
|
Atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr16_-_48692476 | 0.03 |
ENSRNOT00000013118
|
Irf2
|
interferon regulatory factor 2 |
| chr1_+_266844480 | 0.03 |
ENSRNOT00000027482
|
Taf5
|
TATA-box binding protein associated factor 5 |
| chr3_+_60026747 | 0.03 |
ENSRNOT00000081881
|
Scrn3
|
secernin 3 |
| chr4_+_91373942 | 0.02 |
ENSRNOT00000043827
|
Ccser1
|
coiled-coil serine-rich protein 1 |
| chr13_+_90260783 | 0.02 |
ENSRNOT00000050547
|
Cd84
|
CD84 molecule |
| chr2_+_66940057 | 0.02 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr8_+_64610952 | 0.02 |
ENSRNOT00000015963
|
Myo9a
|
myosin IXA |
| chr1_-_197801634 | 0.02 |
ENSRNOT00000090200
|
Nfatc2ip
|
nuclear factor of activated T-cells 2 interacting protein |
| chr5_-_113880911 | 0.02 |
ENSRNOT00000029441
|
Eqtn
|
equatorin |
| chr3_+_158997834 | 0.02 |
ENSRNOT00000000211
|
LOC108348048
|
von Willebrand factor A domain-containing protein 5A |
| chrX_+_84064427 | 0.02 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr3_-_64543100 | 0.02 |
ENSRNOT00000025803
|
Zfp385b
|
zinc finger protein 385B |
| chr13_-_74923402 | 0.02 |
ENSRNOT00000033324
|
Rasal2
|
RAS protein activator like 2 |
| chr1_+_256226179 | 0.02 |
ENSRNOT00000054748
|
Exoc6
|
exocyst complex component 6 |
| chr16_-_7412150 | 0.02 |
ENSRNOT00000047252
|
RGD1564325
|
similar to ribosomal protein S24 |
| chr16_+_71242470 | 0.02 |
ENSRNOT00000030489
|
Letm2
|
leucine zipper and EF-hand containing transmembrane protein 2 |
| chr10_-_91661558 | 0.02 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr8_+_96551245 | 0.02 |
ENSRNOT00000039850
|
Bcl2a1
|
BCL2-related protein A1 |
| chr20_+_3167079 | 0.02 |
ENSRNOT00000001035
|
RT1-N3
|
RT1 class Ib, locus N3 |
| chr8_+_31497639 | 0.02 |
ENSRNOT00000020041
|
Snx19
|
sorting nexin 19 |
| chr3_-_153321352 | 0.02 |
ENSRNOT00000064824
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr3_-_60166013 | 0.02 |
ENSRNOT00000024922
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr3_-_150960030 | 0.02 |
ENSRNOT00000024714
|
Ncoa6
|
nuclear receptor coactivator 6 |
| chr19_-_11669578 | 0.02 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr14_-_45863877 | 0.02 |
ENSRNOT00000002977
|
Pgm2
|
phosphoglucomutase 2 |
| chrX_+_88298266 | 0.02 |
ENSRNOT00000041508
|
AABR07039936.1
|
|
| chr2_+_196139755 | 0.02 |
ENSRNOT00000040501
|
Pi4kb
|
phosphatidylinositol 4-kinase beta |
| chr3_+_47995959 | 0.01 |
ENSRNOT00000082775
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr2_-_43067956 | 0.01 |
ENSRNOT00000090616
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chrX_+_77076106 | 0.01 |
ENSRNOT00000091527
ENSRNOT00000089381 |
Atp7a
|
ATPase copper transporting alpha |
| chr7_-_70319346 | 0.01 |
ENSRNOT00000071543
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr8_+_42443442 | 0.01 |
ENSRNOT00000015986
|
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr1_-_197821936 | 0.01 |
ENSRNOT00000055027
|
Cd19
|
CD19 molecule |
| chr2_-_53313884 | 0.01 |
ENSRNOT00000046951
|
Ghr
|
growth hormone receptor |
| chr4_+_182483194 | 0.01 |
ENSRNOT00000002528
|
Far2
|
fatty acyl CoA reductase 2 |
| chr14_-_62595854 | 0.01 |
ENSRNOT00000003024
|
Yipf7
|
Yip1 domain family, member 7 |
| chr6_+_73358112 | 0.01 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr4_+_34237762 | 0.01 |
ENSRNOT00000084163
|
Mios
|
meiosis regulator for oocyte development |
| chr14_+_83889089 | 0.01 |
ENSRNOT00000078980
|
Morc2
|
MORC family CW-type zinc finger 2 |
| chr8_-_47094352 | 0.01 |
ENSRNOT00000048347
|
Grik4
|
glutamate ionotropic receptor kainate type subunit 4 |
| chrX_+_44907521 | 0.01 |
ENSRNOT00000004901
|
Tbl1x
|
transducin (beta)-like 1 X-linked |
| chr5_+_135354700 | 0.01 |
ENSRNOT00000021850
|
Ipp
|
intracisternal A particle-promoted polypeptide |
| chr13_-_84759098 | 0.01 |
ENSRNOT00000058683
|
LOC685351
|
similar to flavin containing monooxygenase 5 |
| chr2_+_248398917 | 0.01 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr20_+_3146856 | 0.01 |
ENSRNOT00000050159
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr3_+_151335292 | 0.01 |
ENSRNOT00000073642
|
Mmp24
|
matrix metallopeptidase 24 |
| chr7_-_15852930 | 0.01 |
ENSRNOT00000009270
|
LOC691422
|
similar to zinc finger protein 101 |
| chr13_+_110372164 | 0.01 |
ENSRNOT00000049634
|
RGD1560186
|
similar to ribosomal protein L37 |
| chr12_+_2140203 | 0.01 |
ENSRNOT00000084906
|
Camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr4_+_162493908 | 0.01 |
ENSRNOT00000072064
|
Clec2d
|
C-type lectin domain family 2, member D |
| chr6_+_1516158 | 0.01 |
ENSRNOT00000062031
|
LOC103692531
|
uncharacterized LOC103692531 |
| chr18_-_72649915 | 0.01 |
ENSRNOT00000072205
|
AABR07032582.1
|
|
| chr16_+_54332660 | 0.01 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr11_+_85508300 | 0.01 |
ENSRNOT00000038646
|
AABR07034730.3
|
|
| chr4_-_176909075 | 0.01 |
ENSRNOT00000067489
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chr3_-_13896838 | 0.01 |
ENSRNOT00000025399
|
Fbxw2
|
F-box and WD repeat domain containing 2 |
| chr3_-_102151489 | 0.00 |
ENSRNOT00000006349
|
Ano3
|
anoctamin 3 |
| chr13_+_79269973 | 0.00 |
ENSRNOT00000003969
|
Tnfsf4
|
tumor necrosis factor superfamily member 4 |
| chr1_-_7443863 | 0.00 |
ENSRNOT00000088558
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr7_-_101138373 | 0.00 |
ENSRNOT00000043257
|
LOC500877
|
Ab1-152 |
| chr9_+_27408333 | 0.00 |
ENSRNOT00000088416
|
Gsta3
|
glutathione S-transferase alpha 3 |
| chr9_+_113534326 | 0.00 |
ENSRNOT00000066793
|
Ppp4r1
|
protein phosphatase 4, regulatory subunit 1 |
| chr8_-_32431987 | 0.00 |
ENSRNOT00000048460
|
RGD1563958
|
similar to 60S ribosomal protein L32 |
| chr6_+_55465782 | 0.00 |
ENSRNOT00000006682
|
Agr2
|
anterior gradient 2, protein disulphide isomerase family member |
| chr17_-_15566332 | 0.00 |
ENSRNOT00000093743
|
Ecm2
|
extracellular matrix protein 2 |
| chr1_-_134871167 | 0.00 |
ENSRNOT00000076300
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr5_-_73986373 | 0.00 |
ENSRNOT00000064043
ENSRNOT00000032299 |
Tmem245
|
transmembrane protein 245 |
| chr19_+_39126589 | 0.00 |
ENSRNOT00000027561
|
Sntb2
|
syntrophin, beta 2 |
| chr18_-_27452420 | 0.00 |
ENSRNOT00000037888
|
Cdc23
|
cell division cycle 23 |
| chr10_-_67401836 | 0.00 |
ENSRNOT00000073071
|
Crlf3
|
cytokine receptor-like factor 3 |
| chr9_-_88534710 | 0.00 |
ENSRNOT00000020880
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr1_+_164705604 | 0.00 |
ENSRNOT00000051450
ENSRNOT00000071384 |
Olr36
|
olfactory receptor 36 |
| chr14_+_21041621 | 0.00 |
ENSRNOT00000086571
|
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr1_+_167944448 | 0.00 |
ENSRNOT00000087667
|
Olr56
|
olfactory receptor 56 |
| chrX_-_37705263 | 0.00 |
ENSRNOT00000043666
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.1 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.0 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |