Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
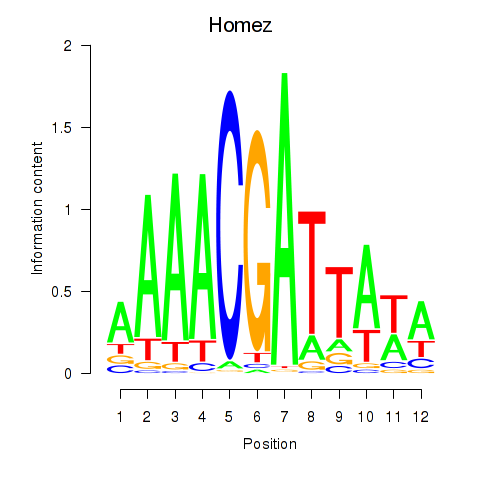
Results for Homez
Z-value: 0.13
Transcription factors associated with Homez
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Homez
|
ENSRNOG00000014887 | homeobox and leucine zipper encoding |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Homez | rn6_v1_chr15_-_33527031_33527031 | 0.82 | 9.2e-02 | Click! |
Activity profile of Homez motif
Sorted Z-values of Homez motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_90948976 | 0.12 |
ENSRNOT00000056877
|
LOC108348111
|
succinate dehydrogenase assembly factor 1, mitochondrial |
| chr10_-_88670430 | 0.10 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr9_+_10305470 | 0.06 |
ENSRNOT00000072592
|
Nrtn
|
neurturin |
| chr8_-_68966108 | 0.06 |
ENSRNOT00000012155
|
Smad6
|
SMAD family member 6 |
| chr10_-_45297385 | 0.05 |
ENSRNOT00000041187
|
Hist3h2bb
|
histone cluster 3 H2B family member b |
| chr10_+_23914894 | 0.05 |
ENSRNOT00000071435
|
Ebf1
|
early B-cell factor 1 |
| chrM_+_7758 | 0.05 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr11_+_84827062 | 0.05 |
ENSRNOT00000058006
|
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr5_-_156734541 | 0.05 |
ENSRNOT00000021036
|
LOC100909857
|
cytidine deaminase-like |
| chr18_-_5314511 | 0.05 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr5_+_139597731 | 0.05 |
ENSRNOT00000072427
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr5_+_147257678 | 0.05 |
ENSRNOT00000066268
|
Rnf19b
|
ring finger protein 19B |
| chr2_-_198339130 | 0.04 |
ENSRNOT00000028767
|
Bola1
|
bolA family member 1 |
| chr8_+_117906014 | 0.04 |
ENSRNOT00000056180
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr1_-_89539210 | 0.04 |
ENSRNOT00000077462
ENSRNOT00000028644 |
Hpn
|
hepsin |
| chr17_+_43633675 | 0.04 |
ENSRNOT00000072119
|
LOC102549173
|
histone H3.2-like |
| chr15_-_26678420 | 0.04 |
ENSRNOT00000041420
|
RGD1310110
|
similar to 3632451O06Rik protein |
| chr19_+_14835822 | 0.04 |
ENSRNOT00000072804
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene |
| chr20_-_2701815 | 0.04 |
ENSRNOT00000061950
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr17_-_78499881 | 0.03 |
ENSRNOT00000079260
|
Fam107b
|
family with sequence similarity 107, member B |
| chr9_-_66019065 | 0.03 |
ENSRNOT00000088729
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chr18_-_69723869 | 0.03 |
ENSRNOT00000000125
|
Elac1
|
elaC ribonuclease Z 1 |
| chr17_+_24242642 | 0.03 |
ENSRNOT00000024323
|
Rnf182
|
ring finger protein 182 |
| chr1_-_67065797 | 0.03 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr1_+_221673590 | 0.03 |
ENSRNOT00000038016
|
Cdc42bpg
|
CDC42 binding protein kinase gamma |
| chr20_+_44680449 | 0.03 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr1_-_7443863 | 0.03 |
ENSRNOT00000088558
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr17_-_14627937 | 0.03 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chrX_-_156155014 | 0.03 |
ENSRNOT00000088637
|
LOC102552182
|
L antigen family member 3-like |
| chr1_+_55219773 | 0.03 |
ENSRNOT00000041610
|
RGD1561667
|
similar to putative protein kinase |
| chr6_-_28994519 | 0.03 |
ENSRNOT00000006668
|
Ubxn2a
|
UBX domain protein 2A |
| chrX_-_45522665 | 0.03 |
ENSRNOT00000030771
|
RGD1562200
|
similar to GS2 gene |
| chr6_+_109043880 | 0.02 |
ENSRNOT00000008730
|
Eif2b2
|
eukaryotic translation initiation factor 2B subunit beta |
| chr13_-_70922245 | 0.02 |
ENSRNOT00000064860
|
Dhx9
|
DExH-box helicase 9 |
| chr2_+_159138758 | 0.02 |
ENSRNOT00000087942
|
AABR07011152.1
|
|
| chr7_-_98735192 | 0.02 |
ENSRNOT00000051430
|
AABR07058017.1
|
|
| chr1_-_100887853 | 0.02 |
ENSRNOT00000027675
|
Med25
|
mediator complex subunit 25 |
| chr8_-_82339937 | 0.02 |
ENSRNOT00000080797
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr12_+_47179664 | 0.02 |
ENSRNOT00000001551
|
Cabp1
|
calcium binding protein 1 |
| chr1_+_105284753 | 0.02 |
ENSRNOT00000041950
|
Slc6a5
|
solute carrier family 6 member 5 |
| chrX_+_71528988 | 0.02 |
ENSRNOT00000075963
|
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr5_-_160742479 | 0.02 |
ENSRNOT00000019447
|
Kazn
|
kazrin, periplakin interacting protein |
| chr8_-_12902262 | 0.02 |
ENSRNOT00000033969
|
Endod1
|
endonuclease domain containing 1 |
| chrX_-_2657155 | 0.02 |
ENSRNOT00000005630
|
Chst7
|
carbohydrate sulfotransferase 7 |
| chr3_-_147819571 | 0.02 |
ENSRNOT00000009840
|
Trib3
|
tribbles pseudokinase 3 |
| chr5_+_39329423 | 0.02 |
ENSRNOT00000010081
|
Gpr63
|
G protein-coupled receptor 63 |
| chr7_-_54855557 | 0.02 |
ENSRNOT00000039475
|
Glipr1l1
|
GLI pathogenesis-related 1 like 1 |
| chr4_+_88328061 | 0.02 |
ENSRNOT00000084775
|
Vom1r87
|
vomeronasal 1 receptor 87 |
| chr10_+_64174931 | 0.02 |
ENSRNOT00000035948
|
RGD1565611
|
RGD1565611 |
| chr3_+_8817076 | 0.02 |
ENSRNOT00000078932
|
Lrrc8a
|
leucine rich repeat containing 8 family, member A |
| chr7_+_133457255 | 0.02 |
ENSRNOT00000080621
|
Cntn1
|
contactin 1 |
| chr10_+_80953006 | 0.02 |
ENSRNOT00000035567
|
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr7_-_71048383 | 0.02 |
ENSRNOT00000005693
|
Gpr182
|
G protein-coupled receptor 182 |
| chr17_-_43776460 | 0.02 |
ENSRNOT00000089055
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr18_+_30587872 | 0.02 |
ENSRNOT00000040016
|
Pcdhb21
|
protocadherin beta 21 |
| chr7_-_12326392 | 0.02 |
ENSRNOT00000039728
|
Ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7 |
| chr1_+_197813963 | 0.02 |
ENSRNOT00000045531
|
LOC100361060
|
ribosomal protein L36-like |
| chr3_-_11820549 | 0.02 |
ENSRNOT00000020660
|
Cfap157
|
cilia and flagella associated protein 157 |
| chr3_+_160231914 | 0.02 |
ENSRNOT00000014411
|
Kcnk15
|
potassium two pore domain channel subfamily K member 15 |
| chr2_-_205212681 | 0.02 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr8_+_87211819 | 0.02 |
ENSRNOT00000086093
|
LOC100363289
|
LRRGT00022-like |
| chr5_+_144054452 | 0.02 |
ENSRNOT00000011565
|
Mrps15
|
mitochondrial ribosomal protein S15 |
| chr1_+_185356975 | 0.02 |
ENSRNOT00000086681
|
Plekha7
|
pleckstrin homology domain containing A7 |
| chr12_-_18526250 | 0.02 |
ENSRNOT00000001791
|
Asmt
|
acetylserotonin O-methyltransferase |
| chr5_-_134526089 | 0.02 |
ENSRNOT00000013321
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1 |
| chr5_-_110832175 | 0.02 |
ENSRNOT00000052379
|
Cks1l
|
CDC28 protein kinase regulatory subunit 1-like |
| chr4_+_24617008 | 0.02 |
ENSRNOT00000086607
|
Ewsr1
|
EWS RNA-binding protein 1 |
| chr3_+_37545238 | 0.02 |
ENSRNOT00000070792
|
Tnfaip6
|
TNF alpha induced protein 6 |
| chr14_-_43992587 | 0.02 |
ENSRNOT00000003425
|
Rhoh
|
ras homolog family member H |
| chr9_+_62291809 | 0.02 |
ENSRNOT00000090621
|
Plcl1
|
phospholipase C-like 1 |
| chr4_-_17440506 | 0.02 |
ENSRNOT00000080931
|
Sema3e
|
semaphorin 3E |
| chr13_-_80862963 | 0.02 |
ENSRNOT00000004864
|
Fmo3
|
flavin containing monooxygenase 3 |
| chr3_-_94833406 | 0.02 |
ENSRNOT00000049695
|
RGD1560017
|
similar to Ac2-210 |
| chr5_+_74874306 | 0.02 |
ENSRNOT00000076918
|
Akap2
|
A-kinase anchoring protein 2 |
| chr20_+_26893016 | 0.02 |
ENSRNOT00000082430
|
Dnajc12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chr5_-_174921 | 0.02 |
ENSRNOT00000056239
|
Tipinl1
|
TIMELESS interacting protein like 1 |
| chr3_-_152179193 | 0.02 |
ENSRNOT00000026700
|
Rbm12
|
RNA binding motif protein 12 |
| chr2_-_235831670 | 0.02 |
ENSRNOT00000037393
|
Ostc
|
oligosaccharyltransferase complex non-catalytic subunit |
| chr8_-_46694613 | 0.02 |
ENSRNOT00000084514
|
AC133265.1
|
|
| chr1_+_7101715 | 0.01 |
ENSRNOT00000019994
|
Sf3b5
|
splicing factor 3b, subunit 5 |
| chr8_+_100260049 | 0.01 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr7_-_107768072 | 0.01 |
ENSRNOT00000093189
|
Ndrg1
|
N-myc downstream regulated 1 |
| chr4_+_88271061 | 0.01 |
ENSRNOT00000087374
|
Vom1r86
|
vomeronasal 1 receptor 86 |
| chr6_+_55881387 | 0.01 |
ENSRNOT00000078548
ENSRNOT00000008312 |
Ispd
|
isoprenoid synthase domain containing |
| chr4_+_57378069 | 0.01 |
ENSRNOT00000080173
ENSRNOT00000011851 |
Nrf1
|
nuclear respiratory factor 1 |
| chr18_+_22964210 | 0.01 |
ENSRNOT00000066816
|
Pik3c3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr17_+_77185053 | 0.01 |
ENSRNOT00000091561
|
Optn
|
optineurin |
| chr11_+_67465236 | 0.01 |
ENSRNOT00000042374
|
Stfa2
|
stefin A2 |
| chr14_+_66598259 | 0.01 |
ENSRNOT00000049743
|
Kcnip4
|
potassium voltage-gated channel interacting protein 4 |
| chr6_+_38474804 | 0.01 |
ENSRNOT00000009779
|
Nbas
|
neuroblastoma amplified sequence |
| chr1_+_229416489 | 0.01 |
ENSRNOT00000028617
|
Olr336
|
olfactory receptor 336 |
| chr1_+_101055622 | 0.01 |
ENSRNOT00000079189
|
Nosip
|
nitric oxide synthase interacting protein |
| chr8_+_107826424 | 0.01 |
ENSRNOT00000019769
|
Dbr1
|
debranching RNA lariats 1 |
| chr10_+_76280933 | 0.01 |
ENSRNOT00000051823
|
RGD1304870
|
similar to Tceb2 protein |
| chr8_-_87245951 | 0.01 |
ENSRNOT00000015299
|
Tmem30a
|
transmembrane protein 30A |
| chr4_+_171748273 | 0.01 |
ENSRNOT00000009998
|
Dera
|
deoxyribose-phosphate aldolase |
| chr1_+_85437780 | 0.01 |
ENSRNOT00000093740
|
Supt5h
|
SPT5 homolog, DSIF elongation factor subunit |
| chr1_-_275882444 | 0.01 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr15_-_37831031 | 0.01 |
ENSRNOT00000091562
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr1_+_78015722 | 0.01 |
ENSRNOT00000002043
|
Kptn
|
kaptin (actin binding protein) |
| chr15_+_18941431 | 0.01 |
ENSRNOT00000092092
|
AABR07017236.1
|
|
| chr17_-_3729810 | 0.01 |
ENSRNOT00000072930
|
AABR07026893.1
|
|
| chr3_+_16846412 | 0.01 |
ENSRNOT00000074266
|
AABR07051551.1
|
|
| chrX_-_14972675 | 0.01 |
ENSRNOT00000079664
|
Slc38a5
|
solute carrier family 38, member 5 |
| chrX_-_75224268 | 0.01 |
ENSRNOT00000089809
|
Abcb7
|
ATP binding cassette subfamily B member 7 |
| chr4_-_164051812 | 0.01 |
ENSRNOT00000085719
|
AABR07062183.1
|
|
| chr20_+_8285502 | 0.01 |
ENSRNOT00000081715
|
Rnf8
|
ring finger protein 8 |
| chr12_+_30198822 | 0.01 |
ENSRNOT00000041645
|
Gusb
|
glucuronidase, beta |
| chr9_+_88493593 | 0.01 |
ENSRNOT00000020712
|
LOC102556337
|
mitochondrial fission factor-like |
| chr12_+_25561777 | 0.01 |
ENSRNOT00000002032
|
Rcc1l
|
RCC1 like |
| chr9_-_85528860 | 0.01 |
ENSRNOT00000063834
ENSRNOT00000088963 |
Wdfy1
|
WD repeat and FYVE domain containing 1 |
| chr10_+_35537977 | 0.01 |
ENSRNOT00000071917
|
Rnf130
|
ring finger protein 130 |
| chr1_-_83990588 | 0.01 |
ENSRNOT00000002052
|
Rab4b
|
RAB4B, member RAS oncogene family |
| chr14_-_85951891 | 0.01 |
ENSRNOT00000016207
ENSRNOT00000080848 |
Mrps24
|
mitochondrial ribosomal protein S24 |
| chr7_+_11545024 | 0.01 |
ENSRNOT00000073221
|
Slc39a3
|
solute carrier family 39 member 3 |
| chr2_+_58462949 | 0.01 |
ENSRNOT00000080618
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr5_-_72000318 | 0.01 |
ENSRNOT00000029187
|
AABR07048308.1
|
|
| chr8_-_16486419 | 0.01 |
ENSRNOT00000033223
|
LOC690235
|
similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase-like 1) |
| chr8_-_118187791 | 0.01 |
ENSRNOT00000048764
|
Dhx30
|
DExH-box helicase 30 |
| chr16_+_67350539 | 0.01 |
ENSRNOT00000085659
ENSRNOT00000015879 |
Unc5d
|
unc-5 netrin receptor D |
| chr17_+_9797907 | 0.01 |
ENSRNOT00000021638
|
Lman2
|
lectin, mannose-binding 2 |
| chr11_+_47200029 | 0.01 |
ENSRNOT00000079598
|
Nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr7_+_117304742 | 0.01 |
ENSRNOT00000059599
|
Grina
|
glutamate ionotropic receptor NMDA type subunit associated protein 1 |
| chr12_-_29952196 | 0.01 |
ENSRNOT00000086175
|
Tmem248
|
transmembrane protein 248 |
| chr5_-_106758032 | 0.01 |
ENSRNOT00000033651
|
Hacd4
|
3-hydroxyacyl-CoA dehydratase 4 |
| chr13_-_83202864 | 0.01 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr17_-_63879166 | 0.01 |
ENSRNOT00000049391
ENSRNOT00000045183 |
Zmynd11
|
zinc finger, MYND-type containing 11 |
| chr4_-_165026414 | 0.01 |
ENSRNOT00000071421
|
Klra1
|
killer cell lectin-like receptor, subfamily A, member 1 |
| chr14_+_6221085 | 0.01 |
ENSRNOT00000068215
|
AABR07014259.1
|
|
| chr3_-_22952063 | 0.01 |
ENSRNOT00000083850
|
Psmb7
|
proteasome subunit beta 7 |
| chr16_+_19762938 | 0.01 |
ENSRNOT00000022650
|
Use1
|
unconventional SNARE in the ER 1 |
| chrX_-_29648359 | 0.01 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr2_+_193572053 | 0.01 |
ENSRNOT00000056479
|
AABR07012329.1
|
|
| chr7_+_117456039 | 0.01 |
ENSRNOT00000051754
|
Mroh1
|
maestro heat-like repeat family member 1 |
| chr1_-_188407132 | 0.01 |
ENSRNOT00000073233
|
Gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr13_+_52667969 | 0.01 |
ENSRNOT00000084986
ENSRNOT00000050284 |
Tnnt2
|
troponin T2, cardiac type |
| chr18_+_30890869 | 0.01 |
ENSRNOT00000060466
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr12_+_4310334 | 0.01 |
ENSRNOT00000041244
|
Vom2r60
|
vomeronasal 2 receptor, 60 |
| chr10_+_45297937 | 0.01 |
ENSRNOT00000066367
|
Trim17
|
tripartite motif-containing 17 |
| chr18_-_58097316 | 0.01 |
ENSRNOT00000088527
|
AABR07032265.1
|
|
| chr2_+_196145384 | 0.01 |
ENSRNOT00000080966
|
Pi4kb
|
phosphatidylinositol 4-kinase beta |
| chr17_+_26785029 | 0.01 |
ENSRNOT00000022065
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr14_-_8548310 | 0.01 |
ENSRNOT00000092436
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr1_+_175839108 | 0.01 |
ENSRNOT00000023741
|
Ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr3_-_37854561 | 0.01 |
ENSRNOT00000076095
|
Neb
|
nebulin |
| chr9_+_46962288 | 0.01 |
ENSRNOT00000082146
|
Il1r1
|
interleukin 1 receptor type 1 |
| chr8_-_48692295 | 0.01 |
ENSRNOT00000065456
|
Vps11
|
VPS11 CORVET/HOPS core subunit |
| chr11_+_60383431 | 0.01 |
ENSRNOT00000093295
|
Cd200
|
Cd200 molecule |
| chr8_-_133128290 | 0.01 |
ENSRNOT00000008783
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr1_+_83933942 | 0.01 |
ENSRNOT00000068690
|
Cyp2f4
|
cytochrome P450, family 2, subfamily f, polypeptide 4 |
| chr19_+_426078 | 0.01 |
ENSRNOT00000089516
|
AABR07042629.1
|
|
| chr11_+_9642365 | 0.01 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr7_+_95309928 | 0.01 |
ENSRNOT00000005887
|
Mtbp
|
MDM2 binding protein |
| chr4_+_10748745 | 0.01 |
ENSRNOT00000046142
|
Rsbn1l
|
round spermatid basic protein 1-like |
| chr2_-_257474733 | 0.01 |
ENSRNOT00000016967
ENSRNOT00000066780 |
Nexn
|
nexilin (F actin binding protein) |
| chr14_+_89253373 | 0.01 |
ENSRNOT00000035922
|
LOC688553
|
hypothetical protein LOC688553 |
| chr1_-_67134827 | 0.01 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr2_-_192381716 | 0.01 |
ENSRNOT00000064950
|
AABR07012291.1
|
|
| chr1_+_102017263 | 0.01 |
ENSRNOT00000028680
|
Nomo1
|
nodal modulator 1 |
| chr1_-_255976425 | 0.01 |
ENSRNOT00000085303
|
Ide
|
insulin degrading enzyme |
| chrX_+_105537602 | 0.01 |
ENSRNOT00000029833
|
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr12_-_10307265 | 0.01 |
ENSRNOT00000092627
|
Wasf3
|
WAS protein family, member 3 |
| chr14_+_108826831 | 0.01 |
ENSRNOT00000083146
ENSRNOT00000009421 |
Bcl11a
|
B-cell CLL/lymphoma 11A |
| chr1_+_67025240 | 0.01 |
ENSRNOT00000051024
|
Vom1r47
|
vomeronasal 1 receptor 47 |
| chr18_+_55666027 | 0.01 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chr13_+_48427038 | 0.01 |
ENSRNOT00000009241
|
Ctse
|
cathepsin E |
| chr10_-_35005287 | 0.01 |
ENSRNOT00000032386
|
LOC103689949
|
nedd4 binding protein 3 |
| chr7_-_123287721 | 0.01 |
ENSRNOT00000009152
|
LOC100359574
|
NHP2 non-histone chromosome protein 2-like 1-like |
| chr1_-_164441167 | 0.01 |
ENSRNOT00000023935
|
Rps3
|
ribosomal protein S3 |
| chr18_+_30509393 | 0.01 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr14_-_21299068 | 0.01 |
ENSRNOT00000065778
|
Amtn
|
amelotin |
| chr18_-_64061472 | 0.01 |
ENSRNOT00000022519
|
Fam210a
|
family with sequence similarity 210, member A |
| chr7_-_12405022 | 0.01 |
ENSRNOT00000021648
|
Cirbp
|
cold inducible RNA binding protein |
| chr2_-_238246931 | 0.01 |
ENSRNOT00000086303
|
Gstcd
|
glutathione S-transferase, C-terminal domain containing |
| chr16_+_19097391 | 0.01 |
ENSRNOT00000018018
|
Calr3
|
calreticulin 3 |
| chr14_+_73674753 | 0.01 |
ENSRNOT00000040676
|
RGD1561694
|
similar to High mobility group protein 2 (HMG-2) |
| chr8_+_116325286 | 0.01 |
ENSRNOT00000017461
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr1_-_175657485 | 0.01 |
ENSRNOT00000024561
|
rnf141
|
ring finger protein 141 |
| chr10_-_37099004 | 0.01 |
ENSRNOT00000075465
|
N4bp3
|
Nedd4 binding protein 3 |
| chr10_+_69412017 | 0.01 |
ENSRNOT00000009448
|
Ccl2
|
C-C motif chemokine ligand 2 |
| chr9_+_17216495 | 0.01 |
ENSRNOT00000026331
|
Mad2l1bp
|
MAD2L1 binding protein |
| chr15_+_75067297 | 0.01 |
ENSRNOT00000057931
|
LOC498555
|
similar to 60S acidic ribosomal protein P2 |
| chr6_-_80153231 | 0.01 |
ENSRNOT00000005568
|
Trappc6b
|
trafficking protein particle complex 6B |
| chr3_-_54593306 | 0.01 |
ENSRNOT00000087122
|
Stk39
|
serine threonine kinase 39 |
| chr1_-_62191818 | 0.01 |
ENSRNOT00000033187
|
AABR07001926.1
|
|
| chr5_-_151029233 | 0.01 |
ENSRNOT00000089155
|
Ppp1r8
|
protein phosphatase 1, regulatory subunit 8 |
| chr20_+_5057701 | 0.01 |
ENSRNOT00000001119
|
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C |
| chr9_+_117583610 | 0.01 |
ENSRNOT00000088647
ENSRNOT00000049426 |
Epb41l3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr3_+_79570625 | 0.01 |
ENSRNOT00000078278
ENSRNOT00000009687 |
Fnbp4
|
formin binding protein 4 |
| chr2_-_88553086 | 0.01 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr6_+_93385457 | 0.01 |
ENSRNOT00000010300
|
Actr10
|
actin-related protein 10 homolog |
| chr5_+_164751440 | 0.01 |
ENSRNOT00000010558
|
RGD1305350
|
similar to RIKEN cDNA 2510039O18 |
| chrX_+_19243759 | 0.01 |
ENSRNOT00000004238
|
Mageh1
|
MAGE family member H1 |
| chr20_-_10013190 | 0.01 |
ENSRNOT00000084726
ENSRNOT00000089112 |
Rsph1
|
radial spoke head 1 homolog |
| chr4_+_24612205 | 0.01 |
ENSRNOT00000057382
|
Ewsr1
|
EWS RNA-binding protein 1 |
| chr1_-_69835185 | 0.01 |
ENSRNOT00000020757
|
LOC108349745
|
spindle assembly abnormal protein 6 homolog |
| chr5_-_134638150 | 0.01 |
ENSRNOT00000013480
|
Tex38
|
testis expressed 38 |
| chr14_+_9226125 | 0.01 |
ENSRNOT00000088047
|
Wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr18_+_30496318 | 0.01 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr1_+_264260505 | 0.01 |
ENSRNOT00000018815
|
Wnt8b
|
wingless-type MMTV integration site family, member 8B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Homez
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.0 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.0 | 0.0 | GO:1904722 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0031249 | denatured protein binding(GO:0031249) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0070697 | activin receptor binding(GO:0070697) |