Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
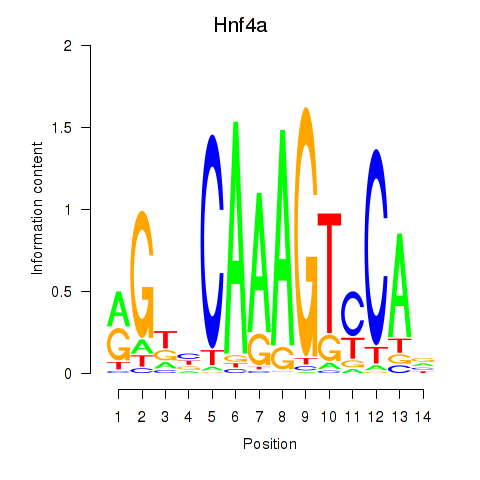
Results for Hnf4a
Z-value: 0.40
Transcription factors associated with Hnf4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hnf4a
|
ENSRNOG00000008895 | hepatocyte nuclear factor 4, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hnf4a | rn6_v1_chr3_+_159902441_159902524 | -0.67 | 2.2e-01 | Click! |
Activity profile of Hnf4a motif
Sorted Z-values of Hnf4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_123638702 | 0.34 |
ENSRNOT00000082473
ENSRNOT00000044470 ENSRNOT00000092017 |
Cyp2d1
|
cytochrome P450, family 2, subfamily d, polypeptide 1 |
| chr4_+_88832178 | 0.12 |
ENSRNOT00000088983
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr10_+_11373346 | 0.10 |
ENSRNOT00000044624
|
LOC108348151
|
coronin-7-like |
| chr2_-_219741886 | 0.10 |
ENSRNOT00000085122
|
Slc35a3
|
solute carrier family 35 member A3 |
| chr2_-_207300854 | 0.09 |
ENSRNOT00000018061
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr6_+_79254339 | 0.09 |
ENSRNOT00000073970
|
Sstr1
|
somatostatin receptor 1 |
| chr1_+_220428481 | 0.08 |
ENSRNOT00000027335
|
LOC108348044
|
ras and Rab interactor 1 |
| chr11_+_87058616 | 0.08 |
ENSRNOT00000002576
ENSRNOT00000082855 |
Prodh1
|
proline dehydrogenase 1 |
| chr4_+_100218661 | 0.08 |
ENSRNOT00000079415
|
Tmem150a
|
transmembrane protein 150A |
| chr11_-_70833577 | 0.08 |
ENSRNOT00000002428
|
Osbpl11
|
oxysterol binding protein-like 11 |
| chr2_+_153803349 | 0.08 |
ENSRNOT00000088565
|
Mme
|
membrane metallo-endopeptidase |
| chr12_-_36398206 | 0.08 |
ENSRNOT00000090944
ENSRNOT00000058492 |
Tmem132b
|
transmembrane protein 132B |
| chr7_-_70467453 | 0.07 |
ENSRNOT00000052288
|
Slc26a10
|
solute carrier family 26, member 10 |
| chr4_+_157659147 | 0.07 |
ENSRNOT00000048379
|
Iffo1
|
intermediate filament family orphan 1 |
| chr8_-_48736506 | 0.07 |
ENSRNOT00000016227
|
Ccdc84
|
coiled-coil domain containing 84 |
| chr8_+_114867062 | 0.07 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr4_+_85551502 | 0.07 |
ENSRNOT00000087191
ENSRNOT00000015692 |
Aqp1
|
aquaporin 1 |
| chr5_+_167141875 | 0.07 |
ENSRNOT00000089314
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr1_+_220335254 | 0.07 |
ENSRNOT00000072261
|
Rin1
|
Ras and Rab interactor 1 |
| chr2_-_47281421 | 0.07 |
ENSRNOT00000086114
|
Itga1
|
integrin subunit alpha 1 |
| chr8_-_48582353 | 0.06 |
ENSRNOT00000011582
|
Pdzd3
|
PDZ domain containing 3 |
| chr1_-_219745654 | 0.06 |
ENSRNOT00000054848
|
LOC689065
|
hypothetical protein LOC689065 |
| chr20_-_2678141 | 0.06 |
ENSRNOT00000072377
ENSRNOT00000083833 |
C4a
|
complement component 4A (Rodgers blood group) |
| chr1_-_175420106 | 0.06 |
ENSRNOT00000013126
ENSRNOT00000077125 |
Sbf2
|
SET binding factor 2 |
| chr9_-_83253458 | 0.06 |
ENSRNOT00000041689
|
Epha4
|
Eph receptor A4 |
| chr15_-_23606634 | 0.06 |
ENSRNOT00000075602
|
Gmfb
|
glia maturation factor, beta |
| chr10_-_73865364 | 0.06 |
ENSRNOT00000005226
|
Rps6kb1
|
ribosomal protein S6 kinase B1 |
| chr20_+_2194709 | 0.06 |
ENSRNOT00000001017
|
Trim15
|
tripartite motif containing 15 |
| chr10_+_64952119 | 0.06 |
ENSRNOT00000012154
|
Pipox
|
pipecolic acid and sarcosine oxidase |
| chr8_-_116361343 | 0.05 |
ENSRNOT00000066296
|
Sema3b
|
semaphorin 3B |
| chr2_-_260884449 | 0.05 |
ENSRNOT00000032874
|
Tyw3
|
tRNA-yW synthesizing protein 3 homolog |
| chr10_+_61432819 | 0.05 |
ENSRNOT00000003687
ENSRNOT00000092478 |
Cluh
|
clustered mitochondria homolog |
| chr7_+_11490852 | 0.05 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chrX_+_14019961 | 0.05 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr5_-_137238354 | 0.05 |
ENSRNOT00000039235
|
Szt2
|
seizure threshold 2 homolog (mouse) |
| chr11_+_69484293 | 0.05 |
ENSRNOT00000049292
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr9_+_81783349 | 0.05 |
ENSRNOT00000021548
|
Cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr13_-_73056765 | 0.05 |
ENSRNOT00000000049
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr10_-_89918427 | 0.05 |
ENSRNOT00000084217
|
Dusp3
|
dual specificity phosphatase 3 |
| chr9_+_10603813 | 0.04 |
ENSRNOT00000073991
ENSRNOT00000074469 ENSRNOT00000079999 |
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr2_+_198965685 | 0.04 |
ENSRNOT00000000107
ENSRNOT00000091578 |
Pdzk1
|
PDZ domain containing 1 |
| chr10_-_31790263 | 0.04 |
ENSRNOT00000059468
|
Timd2
|
T-cell immunoglobulin and mucin domain containing 2 |
| chr17_+_81922329 | 0.04 |
ENSRNOT00000031542
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chrX_+_122938009 | 0.04 |
ENSRNOT00000089266
ENSRNOT00000017550 |
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr7_-_70467915 | 0.04 |
ENSRNOT00000088995
|
Slc26a10
|
solute carrier family 26, member 10 |
| chr5_+_74727494 | 0.04 |
ENSRNOT00000076683
|
Rn50_5_0791.1
|
|
| chr16_+_55152748 | 0.04 |
ENSRNOT00000000121
|
Fgf20
|
fibroblast growth factor 20 |
| chr4_+_157523770 | 0.04 |
ENSRNOT00000055985
ENSRNOT00000023240 |
Zfp384
|
zinc finger protein 384 |
| chr10_+_94170766 | 0.04 |
ENSRNOT00000010627
|
Ace
|
angiotensin I converting enzyme |
| chr1_-_164659992 | 0.04 |
ENSRNOT00000024281
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr20_+_4369394 | 0.04 |
ENSRNOT00000088734
ENSRNOT00000091491 |
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr20_-_2159631 | 0.04 |
ENSRNOT00000061558
|
Trim31
|
tripartite motif-containing 31 |
| chr4_+_157181795 | 0.04 |
ENSRNOT00000017090
|
Lpcat3
|
lysophosphatidylcholine acyltransferase 3 |
| chr1_-_127599257 | 0.04 |
ENSRNOT00000018436
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chr2_-_154542557 | 0.04 |
ENSRNOT00000013392
|
Slc33a1
|
solute carrier family 33 member 1 |
| chr6_-_80107236 | 0.04 |
ENSRNOT00000006369
|
Sec23a
|
Sec23 homolog A, coat complex II component |
| chr7_+_11930135 | 0.04 |
ENSRNOT00000025391
|
Btbd2
|
BTB domain containing 2 |
| chr4_+_157524423 | 0.04 |
ENSRNOT00000036654
|
Zfp384
|
zinc finger protein 384 |
| chr4_-_170740274 | 0.04 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
| chr4_-_10138652 | 0.04 |
ENSRNOT00000017103
|
Lrrc17
|
leucine rich repeat containing 17 |
| chr12_-_11780078 | 0.04 |
ENSRNOT00000076796
|
Zfp498
|
zinc finger protein 498 |
| chr6_+_147876237 | 0.04 |
ENSRNOT00000056649
|
Tmem196
|
transmembrane protein 196 |
| chr7_+_117351648 | 0.04 |
ENSRNOT00000056421
|
Smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr8_-_49271834 | 0.03 |
ENSRNOT00000085022
|
Ube4a
|
ubiquitination factor E4A |
| chr13_+_48790951 | 0.03 |
ENSRNOT00000090233
|
Elk4
|
ELK4, ETS transcription factor |
| chr8_+_104040934 | 0.03 |
ENSRNOT00000081204
|
Tfdp2
|
transcription factor Dp-2 |
| chrX_+_123662477 | 0.03 |
ENSRNOT00000075314
|
Sowahd
|
sosondowah ankyrin repeat domain family member D |
| chr13_+_82479998 | 0.03 |
ENSRNOT00000079872
|
F5
|
coagulation factor V |
| chr10_-_57606171 | 0.03 |
ENSRNOT00000009015
|
Nup88
|
nucleoporin 88 |
| chr4_-_22192474 | 0.03 |
ENSRNOT00000043822
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr6_+_8284878 | 0.03 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr13_+_89586283 | 0.03 |
ENSRNOT00000079355
ENSRNOT00000049873 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr9_-_17163170 | 0.03 |
ENSRNOT00000025921
|
Xpo5
|
exportin 5 |
| chr1_+_88955440 | 0.03 |
ENSRNOT00000091101
|
Prodh2
|
proline dehydrogenase 2 |
| chr1_-_71173216 | 0.03 |
ENSRNOT00000020523
|
Zfp28
|
zinc finger protein 28 |
| chr3_+_172856733 | 0.03 |
ENSRNOT00000009826
|
Edn3
|
endothelin 3 |
| chr3_-_64024205 | 0.03 |
ENSRNOT00000037015
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr5_+_173447784 | 0.03 |
ENSRNOT00000027251
|
Tnfrsf4
|
TNF receptor superfamily member 4 |
| chr15_-_12513931 | 0.03 |
ENSRNOT00000010103
|
Atxn7
|
ataxin 7 |
| chr10_+_104359342 | 0.03 |
ENSRNOT00000006327
|
Tsen54
|
tRNA splicing endonuclease subunit 54 |
| chr1_-_80594136 | 0.03 |
ENSRNOT00000024800
|
Apoc2
|
apolipoprotein C2 |
| chr1_-_265798167 | 0.03 |
ENSRNOT00000079483
|
Ldb1
|
LIM domain binding 1 |
| chr4_-_70934295 | 0.03 |
ENSRNOT00000020616
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr13_-_110784209 | 0.03 |
ENSRNOT00000071698
|
Traf5
|
TNF receptor-associated factor 5 |
| chr15_-_100348760 | 0.03 |
ENSRNOT00000085607
|
AABR07019341.1
|
|
| chr20_+_28572242 | 0.03 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr3_-_175654707 | 0.03 |
ENSRNOT00000080191
|
Rbbp8nl
|
RBBP8 N-terminal like |
| chr14_+_7113544 | 0.03 |
ENSRNOT00000038188
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr7_-_3386522 | 0.03 |
ENSRNOT00000010760
|
Mettl7b
|
methyltransferase like 7B |
| chr3_-_45210474 | 0.03 |
ENSRNOT00000091777
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr3_+_161272385 | 0.03 |
ENSRNOT00000021052
|
Zswim3
|
zinc finger, SWIM-type containing 3 |
| chr10_+_46092754 | 0.03 |
ENSRNOT00000080666
|
Mprip
|
myosin phosphatase Rho interacting protein |
| chr5_+_113725717 | 0.02 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr10_+_89285855 | 0.02 |
ENSRNOT00000028033
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chrX_-_156428593 | 0.02 |
ENSRNOT00000089778
|
Taz
|
tafazzin |
| chr20_-_4316715 | 0.02 |
ENSRNOT00000031704
|
C4b
|
complement C4B (Chido blood group) |
| chr2_-_154542919 | 0.02 |
ENSRNOT00000076880
|
Slc33a1
|
solute carrier family 33 member 1 |
| chr1_+_89491654 | 0.02 |
ENSRNOT00000028632
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chr20_-_4508197 | 0.02 |
ENSRNOT00000086027
ENSRNOT00000000514 |
C4a
|
complement component 4A (Rodgers blood group) |
| chr18_+_59096643 | 0.02 |
ENSRNOT00000025325
|
Wdr7
|
WD repeat domain 7 |
| chr2_-_164634434 | 0.02 |
ENSRNOT00000018402
|
Lxn
|
latexin |
| chr6_-_109935533 | 0.02 |
ENSRNOT00000013516
|
Tgfb3
|
transforming growth factor, beta 3 |
| chr9_-_69953182 | 0.02 |
ENSRNOT00000015852
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr10_+_74886985 | 0.02 |
ENSRNOT00000009943
|
Mtmr4
|
myotubularin related protein 4 |
| chrX_+_135470915 | 0.02 |
ENSRNOT00000009637
|
Slc25a14
|
solute carrier family 25 member 14 |
| chr7_+_2831004 | 0.02 |
ENSRNOT00000029778
|
Rnf41
|
ring finger protein 41 |
| chr10_-_7200499 | 0.02 |
ENSRNOT00000003633
|
Abat
|
4-aminobutyrate aminotransferase |
| chr1_+_14224393 | 0.02 |
ENSRNOT00000016037
|
Perp
|
PERP, TP53 apoptosis effector |
| chr3_+_154507035 | 0.02 |
ENSRNOT00000017265
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr17_-_9762813 | 0.02 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr10_-_15381691 | 0.02 |
ENSRNOT00000027439
|
Rab11fip3
|
RAB11 family interacting protein 3 |
| chr8_+_79489790 | 0.02 |
ENSRNOT00000091722
|
Prtg
|
protogenin |
| chr19_-_55284663 | 0.02 |
ENSRNOT00000018387
|
Snai3
|
snail family transcriptional repressor 3 |
| chr4_+_117215064 | 0.02 |
ENSRNOT00000020909
|
Smyd5
|
SMYD family member 5 |
| chr1_+_85162452 | 0.02 |
ENSRNOT00000093347
|
Pak4
|
p21 (RAC1) activated kinase 4 |
| chr2_+_196304492 | 0.02 |
ENSRNOT00000028640
|
Lysmd1
|
LysM domain containing 1 |
| chrX_+_33884499 | 0.02 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chrX_-_157172068 | 0.02 |
ENSRNOT00000087962
|
Dusp9
|
dual specificity phosphatase 9 |
| chr10_+_62981297 | 0.02 |
ENSRNOT00000031618
|
Efcab5
|
EF-hand calcium binding domain 5 |
| chr20_-_4542073 | 0.02 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr5_-_165259224 | 0.02 |
ENSRNOT00000012692
|
Ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr10_-_82209459 | 0.02 |
ENSRNOT00000004377
|
Spata20
|
spermatogenesis associated 20 |
| chr8_+_31497639 | 0.01 |
ENSRNOT00000020041
|
Snx19
|
sorting nexin 19 |
| chr16_+_26859397 | 0.01 |
ENSRNOT00000044171
|
Msmo1
|
methylsterol monooxygenase 1 |
| chr1_-_7726401 | 0.01 |
ENSRNOT00000021524
|
Pex3
|
peroxisomal biogenesis factor 3 |
| chr7_-_123655896 | 0.01 |
ENSRNOT00000012413
|
Cyp2d2
|
cytochrome P450, family 2, subfamily d, polypeptide 2 |
| chr2_+_62150493 | 0.01 |
ENSRNOT00000080241
|
Zfr
|
zinc finger RNA binding protein |
| chr13_+_110677810 | 0.01 |
ENSRNOT00000006340
|
Slc30a1
|
solute carrier family 30 member 1 |
| chr7_-_120077612 | 0.01 |
ENSRNOT00000011750
|
Lgals2
|
galectin 2 |
| chr2_-_225389120 | 0.01 |
ENSRNOT00000016739
|
Abcd3
|
ATP binding cassette subfamily D member 3 |
| chr10_-_110701137 | 0.01 |
ENSRNOT00000074193
|
Znf750
|
zinc finger protein 750 |
| chr1_-_211956858 | 0.01 |
ENSRNOT00000054886
|
Cfap46
|
cilia and flagella associated protein 46 |
| chr1_+_88955135 | 0.01 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chr3_-_146812989 | 0.01 |
ENSRNOT00000011315
|
Nanp
|
N-acetylneuraminic acid phosphatase |
| chr12_+_47407811 | 0.01 |
ENSRNOT00000001565
|
Hnf1a
|
HNF1 homeobox A |
| chr8_+_63379087 | 0.01 |
ENSRNOT00000012091
ENSRNOT00000012295 |
Nptn
|
neuroplastin |
| chr10_-_15459897 | 0.01 |
ENSRNOT00000027518
|
Decr2
|
2,4-dienoyl-CoA reductase 2 |
| chr15_+_33121273 | 0.01 |
ENSRNOT00000016020
|
Rem2
|
RRAD and GEM like GTPase 2 |
| chr11_+_60140130 | 0.01 |
ENSRNOT00000033595
|
LOC685680
|
similar to TPA-induced transmembrane protein |
| chr8_-_50231357 | 0.01 |
ENSRNOT00000081737
|
Tagln
|
transgelin |
| chr11_-_69201380 | 0.01 |
ENSRNOT00000085618
|
Mylk
|
myosin light chain kinase |
| chr4_-_95970666 | 0.01 |
ENSRNOT00000008826
|
Hpgds
|
hematopoietic prostaglandin D synthase |
| chr5_+_9230984 | 0.01 |
ENSRNOT00000009136
|
Vcpip1
|
valosin containing protein interacting protein 1 |
| chr1_-_219139449 | 0.01 |
ENSRNOT00000023425
ENSRNOT00000089107 |
Tcirg1
|
T-cell immune regulator 1, ATPase H+ transporting V0 subunit A3 |
| chr1_-_224698514 | 0.01 |
ENSRNOT00000024234
|
Slc22a25
|
solute carrier family 22, member 25 |
| chr2_+_58462588 | 0.01 |
ENSRNOT00000083799
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr7_-_15852930 | 0.01 |
ENSRNOT00000009270
|
LOC691422
|
similar to zinc finger protein 101 |
| chrX_-_45284341 | 0.01 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chrX_-_13601069 | 0.01 |
ENSRNOT00000004686
|
Otc
|
ornithine carbamoyltransferase |
| chr19_+_22569999 | 0.01 |
ENSRNOT00000022573
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr9_+_20048121 | 0.01 |
ENSRNOT00000014791
|
Mep1a
|
meprin 1 subunit alpha |
| chr10_-_63274640 | 0.01 |
ENSRNOT00000005129
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr6_+_128050250 | 0.01 |
ENSRNOT00000077517
ENSRNOT00000013961 |
LOC500712
|
Ab1-233 |
| chr12_+_13508933 | 0.01 |
ENSRNOT00000083976
|
Rnf216
|
ring finger protein 216 |
| chr9_-_94201084 | 0.01 |
ENSRNOT00000026190
|
Alpi
|
alkaline phosphatase, intestinal |
| chr2_+_208373154 | 0.01 |
ENSRNOT00000082943
ENSRNOT00000050538 |
LOC100911796
|
adenosine receptor A3-like |
| chr2_+_218834034 | 0.01 |
ENSRNOT00000018367
|
Dph5
|
diphthamide biosynthesis 5 |
| chr5_-_145418992 | 0.01 |
ENSRNOT00000037128
|
Gjb4
|
gap junction protein, beta 4 |
| chr13_+_50434702 | 0.01 |
ENSRNOT00000032244
|
Sox13
|
SRY box 13 |
| chr8_-_130550388 | 0.01 |
ENSRNOT00000026355
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr6_-_39363367 | 0.01 |
ENSRNOT00000088687
ENSRNOT00000065531 |
Fam84a
|
family with sequence similarity 84, member A |
| chr11_+_87381899 | 0.01 |
ENSRNOT00000002554
|
Lztr1
|
leucine-zipper-like transcription regulator 1 |
| chr3_-_120106697 | 0.01 |
ENSRNOT00000020354
|
Prom2
|
prominin 2 |
| chr3_-_153454160 | 0.01 |
ENSRNOT00000010298
|
Ghrh
|
growth hormone releasing hormone |
| chr19_+_49637016 | 0.01 |
ENSRNOT00000016880
|
Bco1
|
beta-carotene oxygenase 1 |
| chr13_+_50434886 | 0.01 |
ENSRNOT00000076857
|
Sox13
|
SRY box 13 |
| chr1_-_127589660 | 0.01 |
ENSRNOT00000084082
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chr16_-_15111585 | 0.01 |
ENSRNOT00000060703
|
LOC683469
|
similar to RNA polymerase II transcription factor SIII subunit A2 (Elongin A2) (EloA2) (Transcription elongation factor B polypeptide 3B) |
| chr3_-_5481144 | 0.01 |
ENSRNOT00000078429
|
Surf4
|
surfeit 4 |
| chr2_+_60180215 | 0.01 |
ENSRNOT00000084624
|
Prlr
|
prolactin receptor |
| chr6_+_33176778 | 0.01 |
ENSRNOT00000046811
ENSRNOT00000007371 |
Apob
|
apolipoprotein B |
| chr2_-_232178533 | 0.01 |
ENSRNOT00000082173
ENSRNOT00000055607 |
Ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr5_+_137257637 | 0.01 |
ENSRNOT00000093001
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr9_-_71788281 | 0.01 |
ENSRNOT00000020078
|
Crygc
|
crystallin, gamma C |
| chr10_+_89286047 | 0.01 |
ENSRNOT00000085831
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr4_+_49056010 | 0.01 |
ENSRNOT00000038566
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr7_-_136853957 | 0.00 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chrX_+_105537602 | 0.00 |
ENSRNOT00000029833
|
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr1_-_82409639 | 0.00 |
ENSRNOT00000031326
|
Erich4
|
glutamate-rich 4 |
| chr10_-_56185857 | 0.00 |
ENSRNOT00000014261
|
Wrap53
|
WD repeat containing, antisense to TP53 |
| chr8_-_103608913 | 0.00 |
ENSRNOT00000013209
|
Pls1
|
plastin 1 |
| chr14_+_77079402 | 0.00 |
ENSRNOT00000042200
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr10_+_99437436 | 0.00 |
ENSRNOT00000006254
|
Kcnj2
|
potassium voltage-gated channel subfamily J member 2 |
| chr1_+_47605415 | 0.00 |
ENSRNOT00000034842
|
Fndc1
|
fibronectin type III domain containing 1 |
| chr2_+_93669765 | 0.00 |
ENSRNOT00000045438
|
Slc10a5
|
solute carrier family 10, member 5 |
| chr1_+_265904566 | 0.00 |
ENSRNOT00000086041
ENSRNOT00000036156 |
Gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chrX_-_64715823 | 0.00 |
ENSRNOT00000076297
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr5_+_137257287 | 0.00 |
ENSRNOT00000037160
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr3_-_5802129 | 0.00 |
ENSRNOT00000009555
|
Sardh
|
sarcosine dehydrogenase |
| chr11_-_46369577 | 0.00 |
ENSRNOT00000049859
ENSRNOT00000047817 ENSRNOT00000002220 ENSRNOT00000002223 ENSRNOT00000063864 ENSRNOT00000070871 ENSRNOT00000072140 |
Abi3bp
|
ABI family member 3 binding protein |
| chr3_-_79743737 | 0.00 |
ENSRNOT00000013584
|
Ptpmt1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr7_-_62162453 | 0.00 |
ENSRNOT00000010720
|
Cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr15_+_12827707 | 0.00 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr6_-_79306443 | 0.00 |
ENSRNOT00000030706
|
Clec14a
|
C-type lectin domain family 14, member A |
| chr15_-_46432965 | 0.00 |
ENSRNOT00000014320
|
Gata4
|
GATA binding protein 4 |
| chr1_+_128199322 | 0.00 |
ENSRNOT00000075131
|
Lysmd4
|
LysM domain containing 4 |
| chr2_-_210873024 | 0.00 |
ENSRNOT00000026051
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr1_+_72692105 | 0.00 |
ENSRNOT00000032439
|
Tmem150b
|
transmembrane protein 150B |
| chr10_-_55965216 | 0.00 |
ENSRNOT00000057058
|
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr6_+_26051396 | 0.00 |
ENSRNOT00000006452
|
Rbks
|
ribokinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hnf4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:1904612 | glucuronoside metabolic process(GO:0019389) response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) |
| 0.0 | 0.3 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0072237 | metanephric proximal tubule development(GO:0072237) |
| 0.0 | 0.1 | GO:2001106 | fasciculation of motor neuron axon(GO:0097156) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.0 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0006554 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0071332 | fructose transport(GO:0015755) cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.0 | GO:1903596 | regulation of gap junction assembly(GO:1903596) |
| 0.0 | 0.0 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.0 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0015168 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.0 | GO:0031711 | bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |