Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
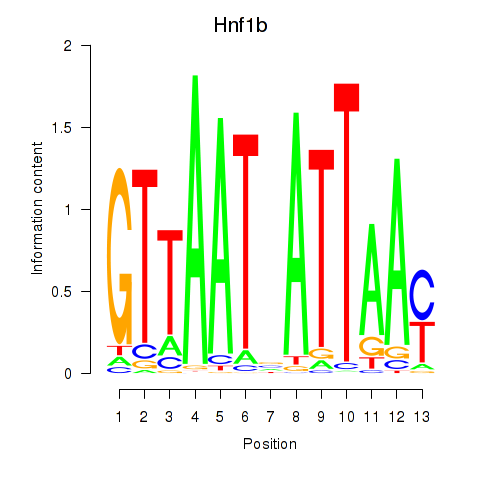
Results for Hnf1b
Z-value: 0.53
Transcription factors associated with Hnf1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hnf1b
|
ENSRNOG00000002598 | HNF1 homeobox B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hnf1b | rn6_v1_chr10_+_71217966_71217966 | 0.93 | 2.4e-02 | Click! |
Activity profile of Hnf1b motif
Sorted Z-values of Hnf1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_32153794 | 0.32 |
ENSRNOT00000005348
|
Tmem27
|
transmembrane protein 27 |
| chr7_-_28711761 | 0.26 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chrX_-_40086870 | 0.25 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr14_+_87448692 | 0.25 |
ENSRNOT00000077177
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr20_+_30690810 | 0.22 |
ENSRNOT00000000687
|
Pcbd1
|
pterin-4 alpha-carbinolamine dehydratase 1 |
| chr8_+_22856539 | 0.19 |
ENSRNOT00000015381
|
Angptl8
|
angiopoietin-like 8 |
| chr10_-_64108554 | 0.18 |
ENSRNOT00000081725
|
Rph3al
|
rabphilin 3A-like (without C2 domains) |
| chr14_-_19132208 | 0.18 |
ENSRNOT00000060535
|
Afm
|
afamin |
| chr4_-_120559078 | 0.17 |
ENSRNOT00000085730
ENSRNOT00000079575 |
Kbtbd12
|
kelch repeat and BTB domain containing 12 |
| chrX_+_33443186 | 0.16 |
ENSRNOT00000005622
|
S100g
|
S100 calcium binding protein G |
| chr14_-_19159923 | 0.16 |
ENSRNOT00000003879
|
Afp
|
alpha-fetoprotein |
| chr3_-_40477754 | 0.15 |
ENSRNOT00000091168
|
AABR07052166.1
|
|
| chr14_+_84231639 | 0.15 |
ENSRNOT00000066362
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr1_+_98414226 | 0.13 |
ENSRNOT00000090785
|
Siglecl1
|
SIGLEC family like 1 |
| chr5_+_10125070 | 0.12 |
ENSRNOT00000085321
|
AABR07046866.1
|
|
| chr14_-_91996774 | 0.12 |
ENSRNOT00000005851
|
Ddc
|
dopa decarboxylase |
| chr2_-_123972356 | 0.12 |
ENSRNOT00000023348
|
Il21
|
interleukin 21 |
| chr2_+_88344527 | 0.11 |
ENSRNOT00000059467
|
RGD1565641
|
RGD1565641 |
| chr8_-_52937972 | 0.11 |
ENSRNOT00000007789
|
Nnmt
|
nicotinamide N-methyltransferase |
| chr13_+_47454591 | 0.11 |
ENSRNOT00000005791
|
LOC498222
|
similar to specifically androgen-regulated protein |
| chr20_-_5123073 | 0.11 |
ENSRNOT00000001126
|
Apom
|
apolipoprotein M |
| chr8_+_58347736 | 0.11 |
ENSRNOT00000080227
ENSRNOT00000066222 |
Slc35f2
|
solute carrier family 35, member F2 |
| chr4_+_172119331 | 0.11 |
ENSRNOT00000010579
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr7_-_119409710 | 0.11 |
ENSRNOT00000008450
|
Ift27
|
intraflagellar transport 27 |
| chr9_+_95274707 | 0.10 |
ENSRNOT00000045163
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr15_-_55277713 | 0.10 |
ENSRNOT00000023037
|
Itm2b
|
integral membrane protein 2B |
| chr10_+_11046024 | 0.09 |
ENSRNOT00000084850
|
Nmral1
|
NmrA like redox sensor 1 |
| chr5_-_12541019 | 0.09 |
ENSRNOT00000091649
|
St18
|
suppression of tumorigenicity 18 |
| chr9_+_95161157 | 0.09 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr19_-_39267928 | 0.09 |
ENSRNOT00000027686
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr2_-_216443518 | 0.09 |
ENSRNOT00000022496
|
Amy1a
|
amylase, alpha 1A |
| chr3_-_127500709 | 0.09 |
ENSRNOT00000006330
|
Hao1
|
hydroxyacid oxidase 1 |
| chr8_-_72204730 | 0.09 |
ENSRNOT00000023810
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr1_+_88955440 | 0.08 |
ENSRNOT00000091101
|
Prodh2
|
proline dehydrogenase 2 |
| chr5_-_79008363 | 0.08 |
ENSRNOT00000010040
|
Kif12
|
kinesin family member 12 |
| chr9_+_95256627 | 0.08 |
ENSRNOT00000025291
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr1_-_101095594 | 0.08 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr14_-_44613904 | 0.08 |
ENSRNOT00000003811
|
Klb
|
klotho beta |
| chr20_-_27117663 | 0.08 |
ENSRNOT00000000434
|
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr18_+_51492196 | 0.08 |
ENSRNOT00000020570
|
Gramd3
|
GRAM domain containing 3 |
| chr5_-_68059933 | 0.07 |
ENSRNOT00000088716
|
Plppr1
|
phospholipid phosphatase related 1 |
| chr11_-_66034573 | 0.07 |
ENSRNOT00000003645
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr3_+_14467330 | 0.07 |
ENSRNOT00000078939
|
Gsn
|
gelsolin |
| chr16_+_25773602 | 0.07 |
ENSRNOT00000047750
|
Gapdh-ps2
|
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 2 |
| chr5_+_138685624 | 0.07 |
ENSRNOT00000011867
|
Guca2a
|
guanylate cyclase activator 2A |
| chr10_+_94471476 | 0.07 |
ENSRNOT00000088697
|
Tcam1
|
testicular cell adhesion molecule 1 |
| chr2_-_170301348 | 0.07 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr3_+_117421604 | 0.07 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr9_+_95295701 | 0.07 |
ENSRNOT00000025045
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr16_+_7758996 | 0.07 |
ENSRNOT00000061063
|
Btd
|
biotinidase |
| chrX_+_35869538 | 0.07 |
ENSRNOT00000058947
|
Ppef1
|
protein phosphatase with EF-hand domain 1 |
| chr7_-_144837583 | 0.06 |
ENSRNOT00000055289
|
Cbx5
|
chromobox 5 |
| chr15_-_41595345 | 0.06 |
ENSRNOT00000019639
|
Sgcg
|
sarcoglycan, gamma |
| chr20_-_25826658 | 0.06 |
ENSRNOT00000057950
ENSRNOT00000084660 |
Ctnna3
|
catenin alpha 3 |
| chr8_-_133128290 | 0.06 |
ENSRNOT00000008783
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr13_-_56958549 | 0.06 |
ENSRNOT00000017293
ENSRNOT00000083912 |
RGD1564614
|
similar to complement factor H-related protein |
| chr4_-_51199570 | 0.06 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr1_+_88955135 | 0.06 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chr18_+_28409955 | 0.06 |
ENSRNOT00000026989
|
Paip2
|
poly(A) binding protein interacting protein 2 |
| chr1_-_248898607 | 0.06 |
ENSRNOT00000074393
|
NEWGENE_1307313
|
dickkopf WNT signaling pathway inhibitor 1 |
| chrX_+_11084317 | 0.06 |
ENSRNOT00000031808
ENSRNOT00000093745 |
RGD1565685
|
similar to RIKEN cDNA 1810030O07 |
| chr3_+_159936856 | 0.06 |
ENSRNOT00000078703
|
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr1_-_15374850 | 0.06 |
ENSRNOT00000016728
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chrX_-_83864150 | 0.06 |
ENSRNOT00000073734
|
Obp1f
|
odorant binding protein I f |
| chr11_-_17684903 | 0.06 |
ENSRNOT00000051213
|
Tmprss15
|
transmembrane protease, serine 15 |
| chr5_+_33097654 | 0.06 |
ENSRNOT00000008087
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr1_+_105284753 | 0.06 |
ENSRNOT00000041950
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr4_-_164453171 | 0.06 |
ENSRNOT00000077539
ENSRNOT00000083610 ENSRNOT00000079975 |
Ly49s6
|
Ly49 stimulatory receptor 6 |
| chr3_+_159902441 | 0.06 |
ENSRNOT00000089893
ENSRNOT00000011978 |
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr7_+_76059386 | 0.06 |
ENSRNOT00000009337
|
Grhl2
|
grainyhead-like transcription factor 2 |
| chr13_-_53870428 | 0.06 |
ENSRNOT00000000812
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr1_-_167893061 | 0.06 |
ENSRNOT00000049401
|
Olr60
|
olfactory receptor 60 |
| chr8_-_127900463 | 0.06 |
ENSRNOT00000078303
|
Slc22a13
|
solute carrier family 22 member 13 |
| chr2_-_158156444 | 0.06 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr18_-_85980833 | 0.05 |
ENSRNOT00000058179
|
Socs6
|
suppressor of cytokine signaling 6 |
| chrX_-_45284341 | 0.05 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chr17_+_53988541 | 0.05 |
ENSRNOT00000079433
|
Tbce
|
tubulin folding cofactor E |
| chr12_-_21746236 | 0.05 |
ENSRNOT00000001869
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr15_-_37831031 | 0.05 |
ENSRNOT00000091562
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr4_+_147333056 | 0.05 |
ENSRNOT00000012137
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
| chr2_-_88553086 | 0.05 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr2_-_166682325 | 0.05 |
ENSRNOT00000091198
ENSRNOT00000012422 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr4_-_59014416 | 0.05 |
ENSRNOT00000049811
|
RGD1561636
|
similar to 60S ribosomal protein L38 |
| chr17_-_69711689 | 0.05 |
ENSRNOT00000041925
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr14_+_77067503 | 0.05 |
ENSRNOT00000085275
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr2_-_88660449 | 0.05 |
ENSRNOT00000051741
|
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr3_+_146980923 | 0.05 |
ENSRNOT00000011654
|
Nsfl1c
|
NSFL1 cofactor |
| chr4_+_82776746 | 0.05 |
ENSRNOT00000011371
|
Tax1bp1
|
Tax1 binding protein 1 |
| chr18_+_57970967 | 0.05 |
ENSRNOT00000039466
|
Spink10
|
serine peptidase inhibitor, Kazal type 10 |
| chr2_-_53718143 | 0.05 |
ENSRNOT00000049363
|
AABR07008350.1
|
|
| chr3_-_9807328 | 0.05 |
ENSRNOT00000064660
|
Tor1a
|
torsin family 1, member A |
| chr4_-_121565212 | 0.05 |
ENSRNOT00000088753
|
Chchd6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr7_-_50034932 | 0.05 |
ENSRNOT00000081885
|
Ptprq
|
protein tyrosine phosphatase, receptor type, Q |
| chr5_+_33784715 | 0.04 |
ENSRNOT00000035685
|
Slc7a13
|
solute carrier family 7 member 13 |
| chr3_-_14229067 | 0.04 |
ENSRNOT00000025534
ENSRNOT00000092865 |
C5
|
complement C5 |
| chr13_+_90514336 | 0.04 |
ENSRNOT00000088996
ENSRNOT00000085377 |
Pex19
|
peroxisomal biogenesis factor 19 |
| chr1_+_101055622 | 0.04 |
ENSRNOT00000079189
|
Nosip
|
nitric oxide synthase interacting protein |
| chr2_+_189922996 | 0.04 |
ENSRNOT00000016499
|
S100a16
|
S100 calcium binding protein A16 |
| chrX_-_46653165 | 0.04 |
ENSRNOT00000058456
|
AABR07038349.1
|
|
| chr10_+_89286047 | 0.04 |
ENSRNOT00000085831
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr15_+_56756661 | 0.04 |
ENSRNOT00000013324
|
Esd
|
esterase D |
| chr1_-_93949187 | 0.04 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr19_-_28296640 | 0.04 |
ENSRNOT00000041115
ENSRNOT00000042781 |
AABR07043429.1
|
|
| chr8_-_77398156 | 0.04 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr1_+_277068761 | 0.04 |
ENSRNOT00000044183
ENSRNOT00000022382 |
Habp2
|
hyaluronan binding protein 2 |
| chr2_-_182035032 | 0.04 |
ENSRNOT00000009813
|
Fgb
|
fibrinogen beta chain |
| chr3_+_80081647 | 0.04 |
ENSRNOT00000057161
|
Arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chrX_+_112270986 | 0.04 |
ENSRNOT00000091441
ENSRNOT00000082652 |
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr17_-_43543172 | 0.04 |
ENSRNOT00000080684
ENSRNOT00000029626 ENSRNOT00000082719 |
Slc17a3
|
solute carrier family 17 member 3 |
| chr8_+_40001030 | 0.04 |
ENSRNOT00000083778
|
Esam
|
endothelial cell adhesion molecule |
| chr5_-_126053726 | 0.04 |
ENSRNOT00000008535
|
Pcsk9
|
proprotein convertase subtilisin/kexin type 9 |
| chr12_-_22006533 | 0.04 |
ENSRNOT00000059593
|
LOC100362783
|
Uncharacterized protein C7orf61 homolog |
| chr6_-_127319362 | 0.04 |
ENSRNOT00000012256
|
Ddx24
|
DEAD-box helicase 24 |
| chr1_+_83965608 | 0.04 |
ENSRNOT00000079995
|
Cyp2t1
|
cytochrome P450, family 2, subfamily t, polypeptide 1 |
| chr12_-_39667849 | 0.04 |
ENSRNOT00000011499
|
Arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr6_+_100337226 | 0.04 |
ENSRNOT00000011220
|
Fut8
|
fucosyltransferase 8 |
| chr1_-_199341302 | 0.04 |
ENSRNOT00000073596
|
Vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr3_+_31802999 | 0.04 |
ENSRNOT00000041305
|
AABR07051996.1
|
|
| chr5_-_131860637 | 0.04 |
ENSRNOT00000064569
ENSRNOT00000080242 |
Slc5a9
|
solute carrier family 5 member 9 |
| chr4_-_30380119 | 0.04 |
ENSRNOT00000036460
|
Pon2
|
paraoxonase 2 |
| chr10_+_47081834 | 0.03 |
ENSRNOT00000065294
|
Dhrs7b
|
dehydrogenase/reductase 7B |
| chr13_-_32427177 | 0.03 |
ENSRNOT00000044628
|
Cdh19
|
cadherin 19 |
| chr7_-_30105132 | 0.03 |
ENSRNOT00000091227
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr15_+_104026601 | 0.03 |
ENSRNOT00000013520
ENSRNOT00000083269 ENSRNOT00000093403 |
Cldn10
|
claudin 10 |
| chr6_-_33738825 | 0.03 |
ENSRNOT00000039492
|
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr1_+_189328246 | 0.03 |
ENSRNOT00000084260
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr19_-_24614019 | 0.03 |
ENSRNOT00000005124
|
Scoc
|
short coiled-coil protein |
| chr3_-_171134655 | 0.03 |
ENSRNOT00000028960
|
RGD1561252
|
similar to Ubiquitin carboxyl-terminal hydrolase isozyme L3 (Ubiquitin thiolesterase L3) |
| chr5_-_124403195 | 0.03 |
ENSRNOT00000067850
|
C8a
|
complement C8 alpha chain |
| chr13_-_102756174 | 0.03 |
ENSRNOT00000029439
|
Marc2
|
mitochondrial amidoxime reducing component 2 |
| chr17_-_43584152 | 0.03 |
ENSRNOT00000023241
|
Slc17a2
|
solute carrier family 17, member 2 |
| chr1_+_74975364 | 0.03 |
ENSRNOT00000047147
ENSRNOT00000090708 |
Vom2r31
|
vomeronasal 2 receptor, 31 |
| chr2_-_88763733 | 0.03 |
ENSRNOT00000059424
|
LOC688389
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr19_+_12097632 | 0.03 |
ENSRNOT00000049452
|
Ces5a
|
carboxylesterase 5A |
| chr8_+_71216178 | 0.03 |
ENSRNOT00000021372
|
Oaz2
|
ornithine decarboxylase antizyme 2 |
| chr2_-_100372252 | 0.03 |
ENSRNOT00000011890
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr8_-_127912860 | 0.03 |
ENSRNOT00000040498
|
LOC685081
|
similar to solute carrier family 22 (organic cation transporter), member 13 |
| chr3_+_91191837 | 0.03 |
ENSRNOT00000006097
|
Rag2
|
recombination activating 2 |
| chr15_-_61772516 | 0.03 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chrX_+_131381134 | 0.03 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chrX_-_64702441 | 0.03 |
ENSRNOT00000051132
|
Amer1
|
APC membrane recruitment protein 1 |
| chr10_-_71491743 | 0.03 |
ENSRNOT00000038955
|
LOC102552988
|
uncharacterized LOC102552988 |
| chr4_+_87293871 | 0.03 |
ENSRNOT00000090943
|
AABR07060628.1
|
|
| chr6_-_102196138 | 0.03 |
ENSRNOT00000014132
|
Tmem229b
|
transmembrane protein 229B |
| chr13_-_47377703 | 0.03 |
ENSRNOT00000005461
|
C4bpa
|
complement component 4 binding protein, alpha |
| chr5_-_128730446 | 0.03 |
ENSRNOT00000089441
|
Osbpl9
|
oxysterol binding protein-like 9 |
| chr19_+_568287 | 0.03 |
ENSRNOT00000016419
|
Cdh16
|
cadherin 16 |
| chr14_+_22724070 | 0.03 |
ENSRNOT00000089471
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr1_+_250426158 | 0.03 |
ENSRNOT00000067643
|
A1cf
|
APOBEC1 complementation factor |
| chr13_+_96303703 | 0.03 |
ENSRNOT00000084718
ENSRNOT00000029723 |
Efcab2
|
EF-hand calcium binding domain 2 |
| chr11_-_24199968 | 0.03 |
ENSRNOT00000074032
|
Mrpl39
|
mitochondrial ribosomal protein L39 |
| chr7_-_97071968 | 0.03 |
ENSRNOT00000006596
|
Slc22a22
|
solute carrier family 22 (organic cation transporter), member 22 |
| chr20_+_47596575 | 0.03 |
ENSRNOT00000087230
ENSRNOT00000064905 |
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr6_-_44363915 | 0.03 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr14_-_19191863 | 0.03 |
ENSRNOT00000003921
|
Alb
|
albumin |
| chr8_+_100260049 | 0.03 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr1_-_266142538 | 0.03 |
ENSRNOT00000026792
|
Actr1a
|
ARP1 actin-related protein 1 homolog A, centractin alpha |
| chr7_+_29435444 | 0.03 |
ENSRNOT00000008613
|
Slc5a8
|
solute carrier family 5 member 8 |
| chr10_-_90127600 | 0.02 |
ENSRNOT00000028368
|
Lsm12
|
LSM12 homolog |
| chr10_-_13446135 | 0.02 |
ENSRNOT00000084991
|
Kctd5
|
potassium channel tetramerization domain containing 5 |
| chr2_-_158156150 | 0.02 |
ENSRNOT00000016621
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr6_-_26385761 | 0.02 |
ENSRNOT00000073228
|
Gckr
|
glucokinase regulator |
| chr4_+_173732248 | 0.02 |
ENSRNOT00000041499
|
Pik3c2g
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr4_+_61912210 | 0.02 |
ENSRNOT00000013569
|
Bpgm
|
bisphosphoglycerate mutase |
| chr3_-_2453933 | 0.02 |
ENSRNOT00000014060
|
Slc34a3
|
solute carrier family 34 member 3 |
| chr1_-_228263198 | 0.02 |
ENSRNOT00000028572
|
Olr1874
|
olfactory receptor 1874 |
| chr1_-_48563776 | 0.02 |
ENSRNOT00000023368
|
Plg
|
plasminogen |
| chr2_+_182006242 | 0.02 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr10_+_57125961 | 0.02 |
ENSRNOT00000029230
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr11_-_33003021 | 0.02 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr1_+_189514553 | 0.02 |
ENSRNOT00000020039
|
Acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr1_+_13261876 | 0.02 |
ENSRNOT00000090703
|
Reps1
|
RALBP1 associated Eps domain containing 1 |
| chr4_-_163762434 | 0.02 |
ENSRNOT00000081854
|
Ly49si1
|
immunoreceptor Ly49si1 |
| chr4_-_149988295 | 0.02 |
ENSRNOT00000019649
|
Fxyd4
|
FXYD domain-containing ion transport regulator 4 |
| chr1_-_62558033 | 0.02 |
ENSRNOT00000015520
|
Zfp40
|
zinc finger protein 40 |
| chr4_-_176381477 | 0.02 |
ENSRNOT00000048367
|
Slco1a6
|
solute carrier organic anion transporter family, member 1a6 |
| chr2_-_243407608 | 0.02 |
ENSRNOT00000014631
|
Mttp
|
microsomal triglyceride transfer protein |
| chr1_+_31124825 | 0.02 |
ENSRNOT00000092105
|
AABR07000989.1
|
|
| chr14_+_100135271 | 0.02 |
ENSRNOT00000032965
|
Fbxo48
|
F-box protein 48 |
| chr1_-_99632845 | 0.02 |
ENSRNOT00000049123
|
LOC103691107
|
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4 pseudogene |
| chr20_+_13498926 | 0.02 |
ENSRNOT00000070992
ENSRNOT00000045375 |
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr14_-_19072677 | 0.02 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr6_-_60124274 | 0.02 |
ENSRNOT00000059823
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr12_+_29921443 | 0.02 |
ENSRNOT00000001190
|
Sbds
|
SBDS ribosome assembly guanine nucleotide exchange factor |
| chr16_-_8686131 | 0.02 |
ENSRNOT00000080202
|
Chat
|
choline O-acetyltransferase |
| chr2_+_195684210 | 0.01 |
ENSRNOT00000028316
|
mrpl9
|
mitochondrial ribosomal protein L9 |
| chr2_+_235264219 | 0.01 |
ENSRNOT00000086245
|
Cfi
|
complement factor I |
| chr4_-_155051429 | 0.01 |
ENSRNOT00000020094
|
Klrg1
|
killer cell lectin like receptor G1 |
| chr10_-_12629114 | 0.01 |
ENSRNOT00000072967
|
Olr1374
|
olfactory receptor 1374 |
| chr3_-_150885475 | 0.01 |
ENSRNOT00000034708
ENSRNOT00000038809 |
Pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr20_-_5037022 | 0.01 |
ENSRNOT00000091022
ENSRNOT00000060762 |
Msh5
|
mutS homolog 5 |
| chr7_-_120077612 | 0.01 |
ENSRNOT00000011750
|
Lgals2
|
galectin 2 |
| chr1_-_267463694 | 0.01 |
ENSRNOT00000084851
ENSRNOT00000016645 |
Col17a1
|
collagen type XVII alpha 1 chain |
| chr18_-_67224566 | 0.01 |
ENSRNOT00000064947
|
Dcc
|
DCC netrin 1 receptor |
| chrX_-_142164220 | 0.01 |
ENSRNOT00000064780
|
Fgf13
|
fibroblast growth factor 13 |
| chr2_+_74360622 | 0.01 |
ENSRNOT00000014013
|
Cdh18
|
cadherin 18 |
| chr14_-_80973456 | 0.01 |
ENSRNOT00000013257
|
Hgfac
|
HGF activator |
| chr14_+_80195715 | 0.01 |
ENSRNOT00000010784
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr4_+_68849033 | 0.01 |
ENSRNOT00000016912
|
Mgam
|
maltase-glucoamylase |
| chr7_-_30274984 | 0.01 |
ENSRNOT00000092527
ENSRNOT00000010207 |
Slc17a8
|
solute carrier family 17 member 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hnf1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.3 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.3 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.0 | 0.1 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:1902569 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of establishment of T cell polarity(GO:1903903) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.0 | 0.1 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.2 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.0 | GO:0010034 | response to acetate(GO:0010034) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.2 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.0 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:1904975 | response to bleomycin(GO:1904975) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.2 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 0.1 | GO:0098603 | selenol Se-methyltransferase activity(GO:0098603) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.0 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |