Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
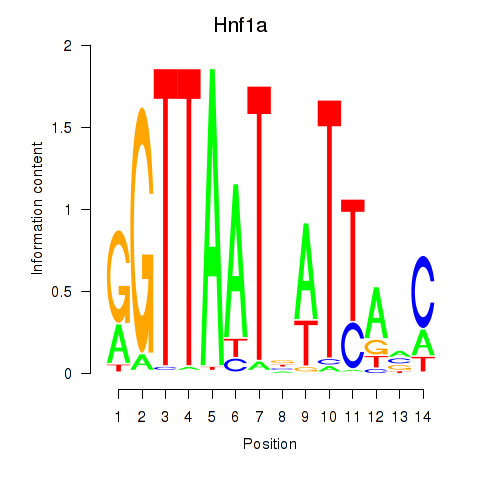
Results for Hnf1a
Z-value: 0.48
Transcription factors associated with Hnf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hnf1a
|
ENSRNOG00000001183 | HNF1 homeobox A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hnf1a | rn6_v1_chr12_+_47407811_47407811 | 0.12 | 8.5e-01 | Click! |
Activity profile of Hnf1a motif
Sorted Z-values of Hnf1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_32153794 | 0.32 |
ENSRNOT00000005348
|
Tmem27
|
transmembrane protein 27 |
| chr14_-_19132208 | 0.30 |
ENSRNOT00000060535
|
Afm
|
afamin |
| chr14_+_84231639 | 0.14 |
ENSRNOT00000066362
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr19_-_39267928 | 0.14 |
ENSRNOT00000027686
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr8_+_22856539 | 0.13 |
ENSRNOT00000015381
|
Angptl8
|
angiopoietin-like 8 |
| chr4_+_66670618 | 0.11 |
ENSRNOT00000010796
|
Tbxas1
|
thromboxane A synthase 1 |
| chr6_-_127534247 | 0.11 |
ENSRNOT00000012500
|
Serpina6
|
serpin family A member 6 |
| chr5_-_68059933 | 0.08 |
ENSRNOT00000088716
|
Plppr1
|
phospholipid phosphatase related 1 |
| chr13_-_50549981 | 0.08 |
ENSRNOT00000003918
ENSRNOT00000080486 |
Golt1a
|
golgi transport 1A |
| chr3_-_54625414 | 0.08 |
ENSRNOT00000037133
|
Stk39
|
serine threonine kinase 39 |
| chr8_-_72204730 | 0.08 |
ENSRNOT00000023810
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr14_-_80973456 | 0.08 |
ENSRNOT00000013257
|
Hgfac
|
HGF activator |
| chr9_+_95295701 | 0.07 |
ENSRNOT00000025045
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr18_+_51492196 | 0.07 |
ENSRNOT00000020570
|
Gramd3
|
GRAM domain containing 3 |
| chr15_-_61772516 | 0.07 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chr20_-_5123073 | 0.07 |
ENSRNOT00000001126
|
Apom
|
apolipoprotein M |
| chr13_-_91776397 | 0.07 |
ENSRNOT00000073147
|
Mptx1
|
mucosal pentraxin 1 |
| chr3_+_159902441 | 0.07 |
ENSRNOT00000089893
ENSRNOT00000011978 |
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr5_-_79008363 | 0.07 |
ENSRNOT00000010040
|
Kif12
|
kinesin family member 12 |
| chr11_-_17684903 | 0.07 |
ENSRNOT00000051213
|
Tmprss15
|
transmembrane protease, serine 15 |
| chr1_+_277068761 | 0.07 |
ENSRNOT00000044183
ENSRNOT00000022382 |
Habp2
|
hyaluronan binding protein 2 |
| chr14_-_19191863 | 0.07 |
ENSRNOT00000003921
|
Alb
|
albumin |
| chr2_-_88553086 | 0.06 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr14_+_87448692 | 0.06 |
ENSRNOT00000077177
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr3_-_14229067 | 0.06 |
ENSRNOT00000025534
ENSRNOT00000092865 |
C5
|
complement C5 |
| chr14_-_19159923 | 0.06 |
ENSRNOT00000003879
|
Afp
|
alpha-fetoprotein |
| chr5_+_33784715 | 0.06 |
ENSRNOT00000035685
|
Slc7a13
|
solute carrier family 7 member 13 |
| chr2_-_88660449 | 0.06 |
ENSRNOT00000051741
|
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr1_-_48563776 | 0.06 |
ENSRNOT00000023368
|
Plg
|
plasminogen |
| chr2_+_56887987 | 0.05 |
ENSRNOT00000041288
|
Gdnf
|
glial cell derived neurotrophic factor |
| chr10_+_57125961 | 0.05 |
ENSRNOT00000029230
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr19_+_12097632 | 0.05 |
ENSRNOT00000049452
|
Ces5a
|
carboxylesterase 5A |
| chr20_-_5037022 | 0.05 |
ENSRNOT00000091022
ENSRNOT00000060762 |
Msh5
|
mutS homolog 5 |
| chr3_-_127500709 | 0.05 |
ENSRNOT00000006330
|
Hao1
|
hydroxyacid oxidase 1 |
| chr2_-_170301348 | 0.05 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr20_+_13498926 | 0.05 |
ENSRNOT00000070992
ENSRNOT00000045375 |
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chrX_+_119390013 | 0.05 |
ENSRNOT00000074269
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr2_-_182035032 | 0.05 |
ENSRNOT00000009813
|
Fgb
|
fibrinogen beta chain |
| chr12_-_39667849 | 0.05 |
ENSRNOT00000011499
|
Arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr13_-_53870428 | 0.05 |
ENSRNOT00000000812
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr11_+_64882288 | 0.04 |
ENSRNOT00000077727
|
Pla1a
|
phospholipase A1 member A |
| chr13_-_47377703 | 0.04 |
ENSRNOT00000005461
|
C4bpa
|
complement component 4 binding protein, alpha |
| chr20_-_27117663 | 0.04 |
ENSRNOT00000000434
|
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr2_-_158156444 | 0.04 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chrX_+_71272042 | 0.04 |
ENSRNOT00000076034
ENSRNOT00000076816 |
Gjb1
|
gap junction protein, beta 1 |
| chr8_-_77398156 | 0.04 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr6_+_101603319 | 0.04 |
ENSRNOT00000030470
|
Gphn
|
gephyrin |
| chr6_-_26385761 | 0.04 |
ENSRNOT00000073228
|
Gckr
|
glucokinase regulator |
| chr1_+_88955135 | 0.04 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chr2_-_100372252 | 0.04 |
ENSRNOT00000011890
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr10_-_71491743 | 0.04 |
ENSRNOT00000038955
|
LOC102552988
|
uncharacterized LOC102552988 |
| chr6_+_107603580 | 0.04 |
ENSRNOT00000036430
|
Dnal1
|
dynein, axonemal, light chain 1 |
| chr3_+_159936856 | 0.04 |
ENSRNOT00000078703
|
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr10_+_89286047 | 0.04 |
ENSRNOT00000085831
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr2_+_182006242 | 0.04 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr1_-_248898607 | 0.04 |
ENSRNOT00000074393
|
NEWGENE_1307313
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr7_+_58814805 | 0.03 |
ENSRNOT00000005909
|
Tspan8
|
tetraspanin 8 |
| chr3_-_80543031 | 0.03 |
ENSRNOT00000022233
|
F2
|
coagulation factor II |
| chr1_+_221470674 | 0.03 |
ENSRNOT00000054835
|
Naaladl1
|
N-acetylated alpha-linked acidic dipeptidase-like 1 |
| chrX_+_112270986 | 0.03 |
ENSRNOT00000091441
ENSRNOT00000082652 |
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr14_+_32257805 | 0.03 |
ENSRNOT00000039626
|
AABR07014804.1
|
|
| chr4_-_51199570 | 0.03 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr3_+_117421604 | 0.03 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr1_+_250426158 | 0.03 |
ENSRNOT00000067643
|
A1cf
|
APOBEC1 complementation factor |
| chr6_-_127620296 | 0.03 |
ENSRNOT00000012577
|
Serpina1
|
serpin family A member 1 |
| chr8_-_127912860 | 0.03 |
ENSRNOT00000040498
|
LOC685081
|
similar to solute carrier family 22 (organic cation transporter), member 13 |
| chr2_-_88763733 | 0.03 |
ENSRNOT00000059424
|
LOC688389
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr3_-_150885475 | 0.03 |
ENSRNOT00000034708
ENSRNOT00000038809 |
Pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr10_-_85974644 | 0.03 |
ENSRNOT00000006098
ENSRNOT00000082974 |
Cacnb1
|
calcium voltage-gated channel auxiliary subunit beta 1 |
| chr2_-_158156150 | 0.02 |
ENSRNOT00000016621
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr20_+_41266566 | 0.02 |
ENSRNOT00000000653
|
Frk
|
fyn-related Src family tyrosine kinase |
| chr14_-_82975263 | 0.02 |
ENSRNOT00000024165
|
Slc5a1
|
solute carrier family 5 member 1 |
| chr19_+_568287 | 0.02 |
ENSRNOT00000016419
|
Cdh16
|
cadherin 16 |
| chr2_-_166682325 | 0.02 |
ENSRNOT00000091198
ENSRNOT00000012422 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr13_+_49005405 | 0.02 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr14_-_86560694 | 0.02 |
ENSRNOT00000075827
|
Npc1l1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr8_+_119382011 | 0.02 |
ENSRNOT00000056060
|
Lrrfip2
|
LRR binding FLII interacting protein 2 |
| chr7_-_68549763 | 0.02 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr1_+_88955440 | 0.02 |
ENSRNOT00000091101
|
Prodh2
|
proline dehydrogenase 2 |
| chr6_-_127508452 | 0.01 |
ENSRNOT00000073709
|
LOC100909524
|
protein Z-dependent protease inhibitor-like |
| chr4_+_68849033 | 0.01 |
ENSRNOT00000016912
|
Mgam
|
maltase-glucoamylase |
| chr20_-_13458110 | 0.01 |
ENSRNOT00000072183
|
Slc5a4
|
solute carrier family 5 member 4 |
| chr6_-_25507073 | 0.01 |
ENSRNOT00000061176
|
Plb1
|
phospholipase B1 |
| chr1_+_214203861 | 0.01 |
ENSRNOT00000023219
|
Rassf7
|
Ras association domain family member 7 |
| chr17_-_69711689 | 0.01 |
ENSRNOT00000041925
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr3_-_11452529 | 0.00 |
ENSRNOT00000020206
|
Slc25a25
|
solute carrier family 25 member 25 |
| chr1_+_213511874 | 0.00 |
ENSRNOT00000078080
ENSRNOT00000016883 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr9_-_88534710 | 0.00 |
ENSRNOT00000020880
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr3_+_80081647 | 0.00 |
ENSRNOT00000057161
|
Arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hnf1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:1902569 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.0 | 0.1 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:1904975 | response to bleomycin(GO:1904975) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.0 | GO:0032304 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) negative regulation of icosanoid secretion(GO:0032304) regulation of metanephros size(GO:0035566) |
| 0.0 | 0.0 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.0 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0072106 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.0 | 0.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0001907 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) response to platinum ion(GO:0070541) |
| 0.0 | 0.0 | GO:1905225 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) response to thyrotropin-releasing hormone(GO:1905225) |
| 0.0 | 0.0 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.0 | 0.0 | GO:0033986 | response to methanol(GO:0033986) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 0.1 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.0 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.0 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |