Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Hmx2
Z-value: 0.12
Transcription factors associated with Hmx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmx2
|
ENSRNOG00000020638 | H6 family homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmx2 | rn6_v1_chr1_+_203515609_203515609 | -0.71 | 1.7e-01 | Click! |
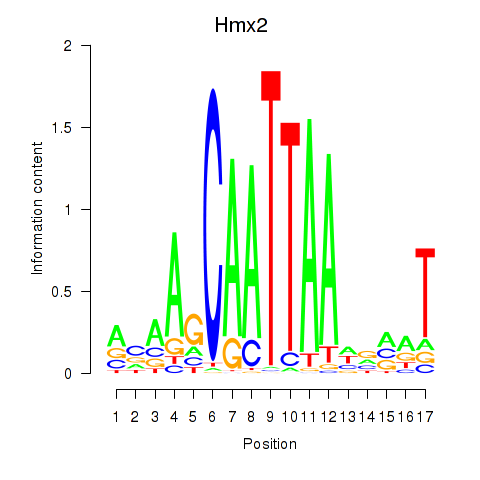
Activity profile of Hmx2 motif
Sorted Z-values of Hmx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_27603307 | 0.07 |
ENSRNOT00000074702
|
AABR07043347.1
|
|
| chr5_-_17061837 | 0.04 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr19_-_21970103 | 0.04 |
ENSRNOT00000074210
|
AABR07043115.1
|
|
| chr5_-_17061361 | 0.04 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr6_+_8886591 | 0.03 |
ENSRNOT00000091510
ENSRNOT00000089174 |
Six3
|
SIX homeobox 3 |
| chr15_-_34198921 | 0.03 |
ENSRNOT00000024991
|
Nrl
|
neural retina leucine zipper |
| chrX_+_77143892 | 0.03 |
ENSRNOT00000085227
|
Atp7a
|
ATPase copper transporting alpha |
| chr4_-_178168690 | 0.03 |
ENSRNOT00000020729
|
Sox5
|
SRY box 5 |
| chr3_-_937102 | 0.03 |
ENSRNOT00000007471
|
Hnmt
|
histamine N-methyltransferase |
| chr11_-_29710849 | 0.03 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr3_-_123698072 | 0.03 |
ENSRNOT00000077819
|
RGD1311739
|
similar to RIKEN cDNA 1700037H04 |
| chr6_+_106153045 | 0.02 |
ENSRNOT00000092065
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chrX_+_115208863 | 0.02 |
ENSRNOT00000072094
|
AABR07040936.1
|
|
| chr15_+_41643541 | 0.02 |
ENSRNOT00000019646
|
Arl11
|
ADP-ribosylation factor like GTPase 11 |
| chr1_+_66898946 | 0.02 |
ENSRNOT00000074063
|
Zfp551
|
zinc finger protein 551 |
| chr10_+_3227160 | 0.02 |
ENSRNOT00000088075
|
Ntan1
|
N-terminal asparagine amidase |
| chr4_+_2053712 | 0.02 |
ENSRNOT00000045086
|
Rnf32
|
ring finger protein 32 |
| chr4_-_171176581 | 0.02 |
ENSRNOT00000030850
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr10_-_103619083 | 0.02 |
ENSRNOT00000029092
|
Cd300e
|
Cd300e molecule |
| chr5_-_78324278 | 0.02 |
ENSRNOT00000082642
ENSRNOT00000048904 |
Wdr31
|
WD repeat domain 31 |
| chr19_-_55284663 | 0.02 |
ENSRNOT00000018387
|
Snai3
|
snail family transcriptional repressor 3 |
| chr2_-_35104963 | 0.02 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr12_-_19592000 | 0.02 |
ENSRNOT00000001855
|
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr1_+_252409268 | 0.01 |
ENSRNOT00000026219
|
Lipm
|
lipase, family member M |
| chr10_-_10767389 | 0.01 |
ENSRNOT00000066754
|
Smim22
|
small integral membrane protein 22 |
| chr1_-_52495582 | 0.01 |
ENSRNOT00000067142
|
Pde10a
|
phosphodiesterase 10A |
| chr20_+_42966140 | 0.01 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr3_+_148327965 | 0.01 |
ENSRNOT00000010851
ENSRNOT00000088481 |
Tpx2
|
TPX2, microtubule nucleation factor |
| chr3_+_44025300 | 0.01 |
ENSRNOT00000006319
|
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr15_-_60752106 | 0.01 |
ENSRNOT00000058148
|
Akap11
|
A-kinase anchoring protein 11 |
| chr17_+_57031766 | 0.01 |
ENSRNOT00000092187
ENSRNOT00000068545 |
Crem
|
cAMP responsive element modulator |
| chr4_-_154044493 | 0.01 |
ENSRNOT00000076318
|
Iqsec3
|
IQ motif and Sec7 domain 3 |
| chr4_+_22859622 | 0.01 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr2_-_138833933 | 0.01 |
ENSRNOT00000013343
|
Pcdh18
|
protocadherin 18 |
| chr15_-_19575914 | 0.01 |
ENSRNOT00000043897
|
AABR07017253.1
|
|
| chr5_+_57657909 | 0.01 |
ENSRNOT00000073017
|
LOC683961
|
similar to ribosomal protein S13 |
| chr2_+_143895982 | 0.01 |
ENSRNOT00000080026
|
Supt20h
|
SPT20 homolog, SAGA complex component |
| chr14_+_37130743 | 0.01 |
ENSRNOT00000031756
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr3_-_165537940 | 0.01 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr2_-_38208719 | 0.01 |
ENSRNOT00000019189
|
Kif2a
|
kinesin family member 2A |
| chr11_-_14304603 | 0.01 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr2_-_189333015 | 0.01 |
ENSRNOT00000076424
ENSRNOT00000076952 |
Hax1
|
HCLS1 associated protein X-1 |
| chr5_+_163773509 | 0.01 |
ENSRNOT00000072167
|
LOC103692519
|
60S ribosomal protein L9 pseudogene |
| chr14_-_34218961 | 0.01 |
ENSRNOT00000072588
|
LOC686911
|
similar to Exocyst complex component 1 (Exocyst complex component Sec3) |
| chr4_-_22425381 | 0.01 |
ENSRNOT00000011166
ENSRNOT00000037712 |
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr1_-_16687817 | 0.01 |
ENSRNOT00000091376
ENSRNOT00000081620 |
Myb
|
MYB proto-oncogene, transcription factor |
| chr3_-_90751055 | 0.01 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr12_+_10254951 | 0.01 |
ENSRNOT00000061129
|
Gpr12
|
G protein-coupled receptor 12 |
| chr18_-_37132223 | 0.01 |
ENSRNOT00000091634
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr5_-_140024176 | 0.01 |
ENSRNOT00000016492
|
Tmco2
|
transmembrane and coiled-coil domains 2 |
| chrX_+_68771100 | 0.01 |
ENSRNOT00000043872
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr17_+_69654170 | 0.01 |
ENSRNOT00000058351
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr2_-_39007976 | 0.01 |
ENSRNOT00000064292
|
Zswim6
|
zinc finger, SWIM-type containing 6 |
| chr6_-_91250138 | 0.01 |
ENSRNOT00000052408
|
LOC100911256
|
ninein-like |
| chr18_+_61563053 | 0.01 |
ENSRNOT00000022845
|
Grp
|
gastrin releasing peptide |
| chrX_+_984798 | 0.01 |
ENSRNOT00000073016
|
Zfp182
|
zinc finger protein 182 |
| chr6_+_107603580 | 0.01 |
ENSRNOT00000036430
|
Dnal1
|
dynein, axonemal, light chain 1 |
| chr12_-_6078411 | 0.01 |
ENSRNOT00000001197
|
Rxfp2
|
relaxin/insulin-like family peptide receptor 2 |
| chr15_+_33885106 | 0.01 |
ENSRNOT00000024483
|
Dhrs2
|
dehydrogenase/reductase 2 |
| chr8_+_106816152 | 0.01 |
ENSRNOT00000047613
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr13_-_55878094 | 0.01 |
ENSRNOT00000014218
|
Lhx9
|
LIM homeobox 9 |
| chr7_-_54778848 | 0.01 |
ENSRNOT00000005399
|
Glipr1
|
GLI pathogenesis-related 1 |
| chr8_+_107229832 | 0.01 |
ENSRNOT00000045821
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr20_+_32450733 | 0.01 |
ENSRNOT00000036449
|
Rsph4a
|
radial spoke head 4 homolog A |
| chr6_+_72891725 | 0.01 |
ENSRNOT00000038074
|
Nubpl
|
nucleotide binding protein-like |
| chr13_-_89343868 | 0.01 |
ENSRNOT00000058497
ENSRNOT00000035400 |
Fcgr2b
|
Fc fragment of IgG receptor IIb |
| chr5_+_154650632 | 0.01 |
ENSRNOT00000072132
|
Zfp46
|
zinc finger protein 46 |
| chr3_+_149524667 | 0.01 |
ENSRNOT00000055345
|
Bpifa2f
|
BPI fold containing family A, member 2F |
| chr2_-_9472384 | 0.01 |
ENSRNOT00000084295
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr2_+_189655702 | 0.01 |
ENSRNOT00000021502
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr17_-_69404323 | 0.01 |
ENSRNOT00000051342
ENSRNOT00000066282 |
Akr1c2
|
aldo-keto reductase family 1, member C2 |
| chr11_+_31893489 | 0.01 |
ENSRNOT00000042750
|
Itsn1
|
intersectin 1 |
| chr2_-_39003552 | 0.01 |
ENSRNOT00000078884
|
Zswim6
|
zinc finger, SWIM-type containing 6 |
| chr7_-_28711761 | 0.01 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chrM_+_9451 | 0.01 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr10_-_104482838 | 0.01 |
ENSRNOT00000007246
|
Recql5
|
RecQ like helicase 5 |
| chr4_-_129619142 | 0.01 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chr20_-_30975472 | 0.01 |
ENSRNOT00000066794
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr18_-_70184337 | 0.01 |
ENSRNOT00000020637
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr7_+_23854846 | 0.01 |
ENSRNOT00000037290
|
Bpifc
|
BPI fold containing family C |
| chr10_-_27427686 | 0.01 |
ENSRNOT00000087475
|
AABR07029410.1
|
|
| chrM_+_7919 | 0.01 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr18_-_69780922 | 0.01 |
ENSRNOT00000079093
|
Me2
|
malic enzyme 2 |
| chr15_-_36798814 | 0.01 |
ENSRNOT00000065764
|
Cenpj
|
centromere protein J |
| chr2_-_170301348 | 0.01 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr2_-_188559882 | 0.01 |
ENSRNOT00000088199
|
Trim46
|
tripartite motif-containing 46 |
| chr18_-_67224566 | 0.01 |
ENSRNOT00000064947
|
Dcc
|
DCC netrin 1 receptor |
| chr4_+_5848722 | 0.01 |
ENSRNOT00000082504
|
AABR07059161.1
|
|
| chr7_+_72599479 | 0.01 |
ENSRNOT00000018963
|
AABR07057460.1
|
|
| chr2_-_199354793 | 0.01 |
ENSRNOT00000023548
|
Bcl9
|
B-cell CLL/lymphoma 9 |
| chr4_+_35279063 | 0.01 |
ENSRNOT00000011833
|
Nxph1
|
neurexophilin 1 |
| chr4_+_156050054 | 0.01 |
ENSRNOT00000039855
|
Clec4a
|
C-type lectin domain family 4, member A |
| chr17_-_48810991 | 0.01 |
ENSRNOT00000067636
|
Vps41
|
VPS41 HOPS complex subunit |
| chr10_-_104522241 | 0.01 |
ENSRNOT00000046010
|
Rps18l1
|
ribosomal protein S18-like 1 |
| chr8_+_126414220 | 0.01 |
ENSRNOT00000092142
|
Azi2
|
5-azacytidine induced 2 |
| chrY_-_1305026 | 0.01 |
ENSRNOT00000092901
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr8_-_126390801 | 0.01 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chr13_+_82574966 | 0.01 |
ENSRNOT00000003844
|
Ccdc181
|
coiled-coil domain containing 181 |
| chr10_+_65552897 | 0.01 |
ENSRNOT00000056217
|
Spag5
|
sperm associated antigen 5 |
| chr1_-_140477796 | 0.01 |
ENSRNOT00000067172
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr9_-_52238564 | 0.01 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr20_+_31234493 | 0.00 |
ENSRNOT00000000675
|
Npffr1
|
neuropeptide FF receptor 1 |
| chrX_-_45284341 | 0.00 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chr7_-_122329443 | 0.00 |
ENSRNOT00000032232
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr10_-_91122018 | 0.00 |
ENSRNOT00000080077
|
Dcakd
|
dephospho-CoA kinase domain containing |
| chr2_+_209433103 | 0.00 |
ENSRNOT00000024036
|
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr20_+_4156858 | 0.00 |
ENSRNOT00000039553
|
Btnl3
|
butyrophilin-like 3 |
| chr12_-_9864791 | 0.00 |
ENSRNOT00000073026
|
Gtf3a
|
general transcription factor III A |
| chr7_+_71709157 | 0.00 |
ENSRNOT00000008211
|
Cpq
|
carboxypeptidase Q |
| chrX_-_23139694 | 0.00 |
ENSRNOT00000033656
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr14_-_21553949 | 0.00 |
ENSRNOT00000031642
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chrX_+_45213228 | 0.00 |
ENSRNOT00000088538
|
LOC103690319
|
odorant-binding protein |
| chr12_+_41486076 | 0.00 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr19_-_9765484 | 0.00 |
ENSRNOT00000016626
|
Setd6
|
SET domain containing 6 |
| chr9_+_8052210 | 0.00 |
ENSRNOT00000073659
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chrX_-_83864150 | 0.00 |
ENSRNOT00000073734
|
Obp1f
|
odorant binding protein I f |
| chr3_-_6054483 | 0.00 |
ENSRNOT00000084491
|
Brd3
|
bromodomain containing 3 |
| chrX_+_124894466 | 0.00 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr5_-_130085838 | 0.00 |
ENSRNOT00000035252
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr8_+_132135187 | 0.00 |
ENSRNOT00000091791
|
Tgm4
|
transglutaminase 4 |
| chr9_+_47536824 | 0.00 |
ENSRNOT00000049349
|
Tmem182
|
transmembrane protein 182 |
| chr10_+_62981297 | 0.00 |
ENSRNOT00000031618
|
Efcab5
|
EF-hand calcium binding domain 5 |
| chr8_+_115546893 | 0.00 |
ENSRNOT00000018932
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr15_-_33576331 | 0.00 |
ENSRNOT00000021851
|
Slc22a17
|
solute carrier family 22, member 17 |
| chr16_+_20657099 | 0.00 |
ENSRNOT00000027053
|
Kxd1
|
KxDL motif containing 1 |
| chr13_+_85465792 | 0.00 |
ENSRNOT00000082991
ENSRNOT00000005277 |
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chrX_+_124631881 | 0.00 |
ENSRNOT00000009329
|
Atp1b4
|
ATPase Na+/K+ transporting family member beta 4 |
| chr13_+_79378733 | 0.00 |
ENSRNOT00000039221
|
Tnfsf18
|
tumor necrosis factor superfamily member 18 |
| chr17_-_9762813 | 0.00 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr5_-_99033107 | 0.00 |
ENSRNOT00000041306
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr1_-_174350196 | 0.00 |
ENSRNOT00000055158
|
LOC499240
|
similar to predicted gene ICRFP703B1614Q5.5 |
| chr5_-_28130803 | 0.00 |
ENSRNOT00000093186
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr16_-_54137660 | 0.00 |
ENSRNOT00000085435
|
Pcm1
|
pericentriolar material 1 |
| chr17_-_77527894 | 0.00 |
ENSRNOT00000032173
|
Bend7
|
BEN domain containing 7 |
| chr2_-_41871858 | 0.00 |
ENSRNOT00000039720
|
Gapt
|
Grb2-binding adaptor protein, transmembrane |
| chr11_+_64472072 | 0.00 |
ENSRNOT00000042756
|
RGD1563835
|
similar to ribosomal protein L27 |
| chr6_+_34071428 | 0.00 |
ENSRNOT00000084212
|
Matn3
|
matrilin 3 |
| chr4_+_14070553 | 0.00 |
ENSRNOT00000077505
|
Cd36
|
CD36 molecule |
| chrX_-_113228265 | 0.00 |
ENSRNOT00000060001
|
RGD1566136
|
similar to 40S ribosomal protein S9 |
| chrX_+_123486989 | 0.00 |
ENSRNOT00000083861
ENSRNOT00000061455 |
Ube2a
|
ubiquitin-conjugating enzyme E2A |
| chr7_-_120027026 | 0.00 |
ENSRNOT00000011215
|
Card10
|
caspase recruitment domain family, member 10 |
| chr16_-_54137485 | 0.00 |
ENSRNOT00000014202
|
Pcm1
|
pericentriolar material 1 |
| chr1_+_249574954 | 0.00 |
ENSRNOT00000074100
|
Cstf2t
|
cleavage stimulation factor subunit 2, tau variant |
| chr7_+_35070284 | 0.00 |
ENSRNOT00000009578
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr4_+_25635765 | 0.00 |
ENSRNOT00000009340
|
Gtpbp10
|
GTP binding protein 10 |
| chr1_-_198706852 | 0.00 |
ENSRNOT00000024074
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr9_+_43834310 | 0.00 |
ENSRNOT00000085720
|
Cnga3
|
cyclic nucleotide gated channel alpha 3 |
| chr3_-_14112851 | 0.00 |
ENSRNOT00000092736
|
C5
|
complement C5 |
| chr18_+_73256085 | 0.00 |
ENSRNOT00000080660
|
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr14_+_108143869 | 0.00 |
ENSRNOT00000090003
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr18_-_58423196 | 0.00 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr11_+_24266345 | 0.00 |
ENSRNOT00000046973
|
Jam2
|
junctional adhesion molecule 2 |
| chr17_+_36691875 | 0.00 |
ENSRNOT00000074314
|
LOC688583
|
similar to High mobility group protein 4 (HMG-4) (High mobility group protein 2a) (HMG-2a) |
| chr17_+_47301511 | 0.00 |
ENSRNOT00000092051
ENSRNOT00000087178 |
Nme8
|
NME/NM23 family member 8 |
| chr5_+_117509354 | 0.00 |
ENSRNOT00000048495
|
LOC100362298
|
ribosomal protein S18-like |
| chr8_-_133002201 | 0.00 |
ENSRNOT00000008772
|
Ccr1
|
C-C motif chemokine receptor 1 |
| chr9_+_47134034 | 0.00 |
ENSRNOT00000020108
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chrX_+_22342308 | 0.00 |
ENSRNOT00000079909
ENSRNOT00000087945 ENSRNOT00000077528 |
Kdm5c
|
lysine demethylase 5C |
| chr2_-_181531978 | 0.00 |
ENSRNOT00000072029
|
Npy2r
|
neuropeptide Y receptor Y2 |
| chr3_+_112228919 | 0.00 |
ENSRNOT00000011761
|
Capn3
|
calpain 3 |
| chr13_-_47397890 | 0.00 |
ENSRNOT00000005505
|
C4bpb
|
complement component 4 binding protein, beta |
| chr4_-_112707270 | 0.00 |
ENSRNOT00000009308
|
Mrpl19
|
mitochondrial ribosomal protein L19 |
| chr5_-_155709215 | 0.00 |
ENSRNOT00000018118
|
Cdc42
|
cell division cycle 42 |
| chr1_-_126211439 | 0.00 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr5_+_163867961 | 0.00 |
ENSRNOT00000081495
|
AABR07050399.1
|
|
| chr7_+_54859326 | 0.00 |
ENSRNOT00000039141
|
Caps2
|
calcyphosine 2 |
| chr1_+_162890475 | 0.00 |
ENSRNOT00000034886
|
Gdpd4
|
glycerophosphodiester phosphodiesterase domain containing 4 |
| chr17_-_76250537 | 0.00 |
ENSRNOT00000092693
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr14_-_10749120 | 0.00 |
ENSRNOT00000003046
|
Cops4
|
COP9 signalosome subunit 4 |
| chr6_-_76552559 | 0.00 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr10_-_40419054 | 0.00 |
ENSRNOT00000039040
|
Ccdc69
|
coiled-coil domain containing 69 |
| chr11_+_68633160 | 0.00 |
ENSRNOT00000063795
|
Sec22a
|
SEC22 homolog A, vesicle trafficking protein |
| chr10_+_15484795 | 0.00 |
ENSRNOT00000079826
|
Tmem8a
|
transmembrane protein 8A |
| chr4_-_138830117 | 0.00 |
ENSRNOT00000008403
|
Il5ra
|
interleukin 5 receptor subunit alpha |
| chr1_-_1116926 | 0.00 |
ENSRNOT00000062027
|
Raet1l
|
retinoic acid early transcript 1L |
| chr6_+_73553210 | 0.00 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr15_+_26887767 | 0.00 |
ENSRNOT00000016674
|
Olr1627
|
olfactory receptor 1627 |
| chr15_+_28023018 | 0.00 |
ENSRNOT00000090272
|
Rnase4
|
ribonuclease A family member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.0 | 0.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.0 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |