Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
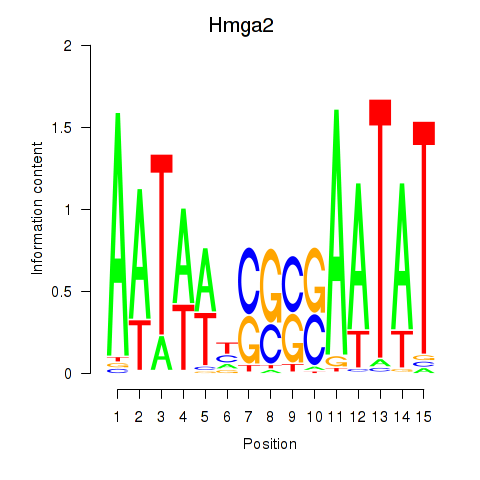
Results for Hmga2
Z-value: 0.41
Transcription factors associated with Hmga2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmga2
|
ENSRNOG00000042460 | high mobility group AT-hook 2 |
Activity profile of Hmga2 motif
Sorted Z-values of Hmga2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_229003961 | 0.19 |
ENSRNOT00000016152
|
Fam111a
|
family with sequence similarity 111, member A |
| chr2_+_184600721 | 0.19 |
ENSRNOT00000075823
|
Gatb
|
glutamyl-tRNA amidotransferase subunit B |
| chr1_-_4560789 | 0.16 |
ENSRNOT00000082415
|
Adgb
|
androglobin |
| chr1_+_168668707 | 0.16 |
ENSRNOT00000015108
|
AC107531.3
|
|
| chr6_+_86785771 | 0.16 |
ENSRNOT00000066702
|
Prpf39
|
pre-mRNA processing factor 39 |
| chr2_-_112368557 | 0.15 |
ENSRNOT00000075017
|
AABR07009787.1
|
|
| chr17_+_48436720 | 0.15 |
ENSRNOT00000074309
|
AABR07027902.1
|
|
| chr15_+_64939527 | 0.15 |
ENSRNOT00000045865
|
AABR07018556.1
|
|
| chr7_-_99808612 | 0.14 |
ENSRNOT00000074953
|
AABR07058091.2
|
|
| chrX_-_70460536 | 0.14 |
ENSRNOT00000076824
|
Pdzd11
|
PDZ domain containing 11 |
| chr6_+_54438436 | 0.14 |
ENSRNOT00000091972
|
AABR07063893.1
|
|
| chr1_+_254531659 | 0.14 |
ENSRNOT00000091377
|
AC080157.1
|
|
| chr1_-_235405831 | 0.14 |
ENSRNOT00000071578
|
AABR07006458.1
|
|
| chr4_-_16654811 | 0.14 |
ENSRNOT00000008637
|
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chrX_+_28072826 | 0.13 |
ENSRNOT00000039796
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr15_-_93765498 | 0.13 |
ENSRNOT00000093297
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr2_+_4942775 | 0.13 |
ENSRNOT00000093548
ENSRNOT00000093741 |
Fam172a
|
family with sequence similarity 172, member A |
| chr2_-_86475096 | 0.13 |
ENSRNOT00000075247
ENSRNOT00000075062 |
LOC100909688
|
zinc finger protein 43-like |
| chr16_+_65256751 | 0.12 |
ENSRNOT00000092012
|
AABR07026137.2
|
|
| chr18_+_14756684 | 0.12 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr3_-_94182714 | 0.11 |
ENSRNOT00000015073
|
LOC100362814
|
hypothetical protein LOC100362814 |
| chr8_+_49418965 | 0.11 |
ENSRNOT00000021819
|
Scn2b
|
sodium voltage-gated channel beta subunit 2 |
| chrX_+_71540895 | 0.11 |
ENSRNOT00000004692
ENSRNOT00000082967 |
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr2_-_231409988 | 0.11 |
ENSRNOT00000080637
ENSRNOT00000037255 ENSRNOT00000074002 |
Ank2
|
ankyrin 2 |
| chr1_-_4011161 | 0.10 |
ENSRNOT00000044778
|
Stxbp5
|
syntaxin binding protein 5 |
| chr12_-_38869346 | 0.10 |
ENSRNOT00000001807
|
Setd1b
|
SET domain containing 1B |
| chr2_-_22798214 | 0.10 |
ENSRNOT00000016135
|
Papd4
|
poly(A) RNA polymerase D4, non-canonical |
| chr15_+_10462749 | 0.10 |
ENSRNOT00000008035
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr2_+_149899836 | 0.10 |
ENSRNOT00000086481
|
RGD1560324
|
similar to hypothetical protein C130079G13 |
| chr12_+_28212333 | 0.10 |
ENSRNOT00000001182
|
Auts2
|
autism susceptibility candidate 2 |
| chr2_+_66940057 | 0.09 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr3_+_101010899 | 0.09 |
ENSRNOT00000073258
|
Lin7c
|
lin-7 homolog C, crumbs cell polarity complex component |
| chr9_+_12346117 | 0.09 |
ENSRNOT00000074599
|
AABR07066677.1
|
|
| chr5_-_166116516 | 0.09 |
ENSRNOT00000079919
ENSRNOT00000080888 |
Kif1b
|
kinesin family member 1B |
| chr18_+_83471342 | 0.09 |
ENSRNOT00000019384
|
Neto1
|
neuropilin and tolloid like 1 |
| chr1_-_66212418 | 0.08 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr5_-_166133491 | 0.08 |
ENSRNOT00000087739
ENSRNOT00000089099 |
Kif1b
|
kinesin family member 1B |
| chr9_+_121831716 | 0.08 |
ENSRNOT00000056250
ENSRNOT00000056251 |
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr8_+_115546712 | 0.08 |
ENSRNOT00000079801
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr1_-_87825333 | 0.08 |
ENSRNOT00000072689
|
Zfp74
|
zinc finger protein 74 |
| chr20_+_28027054 | 0.08 |
ENSRNOT00000071386
ENSRNOT00000001044 |
Ranbp2
|
RAN binding protein 2 |
| chr2_-_231409496 | 0.08 |
ENSRNOT00000055615
ENSRNOT00000015386 ENSRNOT00000068415 |
Ank2
|
ankyrin 2 |
| chr1_+_255710658 | 0.08 |
ENSRNOT00000031545
|
AC094246.1
|
|
| chr5_+_48006823 | 0.08 |
ENSRNOT00000076388
|
Mdn1
|
midasin AAA ATPase 1 |
| chr10_+_89484468 | 0.08 |
ENSRNOT00000035523
|
Tmem106a
|
transmembrane protein 106A |
| chr3_-_158328881 | 0.08 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr6_-_91581262 | 0.07 |
ENSRNOT00000034507
|
AABR07064716.1
|
|
| chr16_+_35573058 | 0.07 |
ENSRNOT00000059580
|
Galntl6
|
polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr14_+_79205466 | 0.07 |
ENSRNOT00000085534
|
Tbc1d14
|
TBC1 domain family, member 14 |
| chr4_+_594882 | 0.07 |
ENSRNOT00000033377
|
Rbm33
|
RNA binding motif protein 33 |
| chr14_+_100311173 | 0.07 |
ENSRNOT00000031275
|
Ppp3r1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chrM_+_9870 | 0.07 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr12_-_17522534 | 0.07 |
ENSRNOT00000047287
|
Sun1
|
Sad1 and UNC84 domain containing 1 |
| chr6_+_59757397 | 0.07 |
ENSRNOT00000047001
|
AABR07064000.1
|
|
| chr14_-_17616170 | 0.07 |
ENSRNOT00000090938
|
Thap6
|
THAP domain containing 6 |
| chr1_+_66898946 | 0.07 |
ENSRNOT00000074063
|
Zfp551
|
zinc finger protein 551 |
| chr3_-_25212049 | 0.07 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr9_-_65387169 | 0.07 |
ENSRNOT00000051843
|
Orc2
|
origin recognition complex, subunit 2 |
| chr16_-_6404578 | 0.07 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr5_+_6373583 | 0.07 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr12_-_51965779 | 0.07 |
ENSRNOT00000056733
|
LOC100362927
|
replication protein A3-like |
| chr17_+_77601877 | 0.07 |
ENSRNOT00000091554
ENSRNOT00000024872 |
Prpf18
|
pre-mRNA processing factor 18 |
| chr8_-_64572850 | 0.07 |
ENSRNOT00000015415
|
Senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chr1_-_88240057 | 0.06 |
ENSRNOT00000073092
|
LOC103690038
|
zinc finger protein 569 |
| chr14_-_92077088 | 0.06 |
ENSRNOT00000085175
|
Grb10
|
growth factor receptor bound protein 10 |
| chr1_+_1545134 | 0.06 |
ENSRNOT00000044523
|
AABR07000156.1
|
|
| chr17_+_5225835 | 0.06 |
ENSRNOT00000022373
|
Zcchc6
|
zinc finger CCHC-type containing 6 |
| chr1_-_48565711 | 0.06 |
ENSRNOT00000023387
|
AABR07001512.1
|
|
| chr2_-_53811134 | 0.06 |
ENSRNOT00000020925
|
Fbxo4
|
F-box protein 4 |
| chr7_+_66717267 | 0.06 |
ENSRNOT00000005654
|
Mon2
|
MON2 homolog, regulator of endosome-to-Golgi trafficking |
| chr18_-_1828606 | 0.06 |
ENSRNOT00000033312
|
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr17_+_43734461 | 0.06 |
ENSRNOT00000072564
|
Hist1h1d
|
histone cluster 1, H1d |
| chr8_-_36374673 | 0.06 |
ENSRNOT00000013763
|
Dcps
|
decapping enzyme, scavenger |
| chr6_+_80108655 | 0.05 |
ENSRNOT00000006137
|
Gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr20_-_4934871 | 0.05 |
ENSRNOT00000071840
|
RT1-CE3
|
RT1 class I, locus CE3 |
| chr15_+_52234563 | 0.05 |
ENSRNOT00000015169
|
Reep4
|
receptor accessory protein 4 |
| chr6_+_43234526 | 0.05 |
ENSRNOT00000086808
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr17_+_57983937 | 0.05 |
ENSRNOT00000022688
|
Wdr37
|
WD repeat domain 37 |
| chr3_-_102151489 | 0.05 |
ENSRNOT00000006349
|
Ano3
|
anoctamin 3 |
| chr10_+_40543288 | 0.05 |
ENSRNOT00000016755
|
Slc36a1
|
solute carrier family 36 member 1 |
| chr4_+_91373942 | 0.05 |
ENSRNOT00000043827
|
Ccser1
|
coiled-coil serine-rich protein 1 |
| chr4_+_31229913 | 0.05 |
ENSRNOT00000077134
ENSRNOT00000087897 |
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr10_-_86144880 | 0.05 |
ENSRNOT00000067901
|
Med1
|
mediator complex subunit 1 |
| chr5_-_163119208 | 0.05 |
ENSRNOT00000085291
ENSRNOT00000022185 |
Vps13d
|
vacuolar protein sorting 13D |
| chr3_-_162035614 | 0.05 |
ENSRNOT00000047890
|
Zfp663
|
zinc finger protein 663 |
| chr13_+_89643881 | 0.05 |
ENSRNOT00000004735
|
B4galt3
|
beta-1,4-galactosyltransferase 3 |
| chr9_-_121931564 | 0.05 |
ENSRNOT00000056243
|
Tyms
|
thymidylate synthetase |
| chrX_-_84768463 | 0.05 |
ENSRNOT00000088570
|
Chm
|
CHM, Rab escort protein 1 |
| chrX_+_124321551 | 0.05 |
ENSRNOT00000074486
|
LOC100910807
|
transcriptional regulator Kaiso-like |
| chr9_-_73958480 | 0.05 |
ENSRNOT00000017838
|
Myl1
|
myosin, light chain 1 |
| chr20_-_19637958 | 0.05 |
ENSRNOT00000074657
|
Slc16a9
|
solute carrier family 16 (monocarboxylic acid transporters), member 9 |
| chrX_+_43881246 | 0.05 |
ENSRNOT00000005153
|
LOC317456
|
hypothetical LOC317456 |
| chr4_+_138269142 | 0.05 |
ENSRNOT00000007788
|
Cntn4
|
contactin 4 |
| chr7_+_35309265 | 0.05 |
ENSRNOT00000080291
|
Tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr20_-_54269525 | 0.05 |
ENSRNOT00000037559
|
Grik2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr4_-_27638676 | 0.05 |
ENSRNOT00000011377
|
RGD1306626
|
similar to RIKEN cDNA 4930500J03 |
| chr4_+_140377565 | 0.05 |
ENSRNOT00000082723
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr1_-_53802658 | 0.05 |
ENSRNOT00000032667
|
Afdn
|
afadin, adherens junction formation factor |
| chr10_+_56610051 | 0.05 |
ENSRNOT00000024348
|
Dvl2
|
dishevelled segment polarity protein 2 |
| chr13_+_56546021 | 0.04 |
ENSRNOT00000016797
|
Aspm
|
abnormal spindle microtubule assembly |
| chr7_-_29378080 | 0.04 |
ENSRNOT00000008317
|
Utp20
|
UTP20 small subunit processome component |
| chr7_+_108613739 | 0.04 |
ENSRNOT00000007564
|
Phf20l1
|
PHD finger protein 20-like 1 |
| chr11_-_74793673 | 0.04 |
ENSRNOT00000002338
ENSRNOT00000002343 |
Opa1
|
OPA1, mitochondrial dynamin like GTPase |
| chr3_-_48831417 | 0.04 |
ENSRNOT00000009920
ENSRNOT00000085246 |
Kcnh7
|
potassium voltage-gated channel subfamily H member 7 |
| chr8_+_117308469 | 0.04 |
ENSRNOT00000080778
|
Qrich1
|
glutamine-rich 1 |
| chr1_-_52495582 | 0.04 |
ENSRNOT00000067142
|
Pde10a
|
phosphodiesterase 10A |
| chrX_+_40258493 | 0.04 |
ENSRNOT00000033010
|
Mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr7_-_59547174 | 0.04 |
ENSRNOT00000085836
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr1_+_61786900 | 0.04 |
ENSRNOT00000090287
|
AABR07001905.1
|
|
| chr5_+_148066062 | 0.04 |
ENSRNOT00000084118
|
AABR07050006.1
|
|
| chr18_-_27550224 | 0.04 |
ENSRNOT00000037368
|
Cdc25c
|
cell division cycle 25C |
| chr5_+_157848206 | 0.04 |
ENSRNOT00000039663
ENSRNOT00000082138 |
Ubr4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr2_+_188139730 | 0.04 |
ENSRNOT00000056789
ENSRNOT00000077698 |
Gon4l
|
gon-4 like |
| chr19_+_37852833 | 0.04 |
ENSRNOT00000006345
|
Nrn1l
|
neuritin 1-like |
| chr2_-_127754648 | 0.04 |
ENSRNOT00000087535
|
Mfsd8
|
major facilitator superfamily domain containing 8 |
| chr20_-_3166569 | 0.03 |
ENSRNOT00000091390
|
AABR07044362.1
|
|
| chr13_+_51317460 | 0.03 |
ENSRNOT00000005845
|
RGD1563962
|
similar to Mss4 protein |
| chr17_-_18421861 | 0.03 |
ENSRNOT00000060500
ENSRNOT00000036560 |
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr1_+_190852985 | 0.03 |
ENSRNOT00000040963
|
Polr3e
|
RNA polymerase III subunit E |
| chr1_+_42169501 | 0.03 |
ENSRNOT00000025477
ENSRNOT00000092791 |
Vip
|
vasoactive intestinal peptide |
| chr2_-_53313884 | 0.03 |
ENSRNOT00000046951
|
Ghr
|
growth hormone receptor |
| chr10_+_3179346 | 0.03 |
ENSRNOT00000082082
|
Rrn3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chrX_+_152885246 | 0.03 |
ENSRNOT00000087074
|
AABR07042326.3
|
|
| chr6_+_113898420 | 0.03 |
ENSRNOT00000064872
|
Nrxn3
|
neurexin 3 |
| chr4_-_28310178 | 0.03 |
ENSRNOT00000084021
|
RGD1563091
|
similar to OEF2 |
| chr5_-_59553416 | 0.03 |
ENSRNOT00000090490
|
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr2_+_186980992 | 0.03 |
ENSRNOT00000020717
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr10_+_3170742 | 0.03 |
ENSRNOT00000093528
ENSRNOT00000004459 |
Rrn3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chr1_-_198117547 | 0.03 |
ENSRNOT00000077226
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr3_+_80670140 | 0.03 |
ENSRNOT00000085614
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chrX_-_157200981 | 0.03 |
ENSRNOT00000087364
|
AABR07071935.1
|
|
| chr3_-_72081079 | 0.03 |
ENSRNOT00000007914
|
Tmx2
|
thioredoxin-related transmembrane protein 2 |
| chr7_-_60860990 | 0.03 |
ENSRNOT00000009511
|
Rap1b
|
RAP1B, member of RAS oncogene family |
| chr3_-_153114520 | 0.03 |
ENSRNOT00000008254
|
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr1_-_38538987 | 0.03 |
ENSRNOT00000090406
ENSRNOT00000071275 |
LOC102551340
|
zinc finger protein 728-like |
| chr3_-_48372583 | 0.03 |
ENSRNOT00000040482
ENSRNOT00000077788 ENSRNOT00000085426 |
Dpp4
|
dipeptidylpeptidase 4 |
| chr11_+_77644541 | 0.03 |
ENSRNOT00000074688
|
Tmem207
|
transmembrane protein 207 |
| chr1_-_31055453 | 0.03 |
ENSRNOT00000031083
|
Soga3
|
SOGA family member 3 |
| chr2_+_243425007 | 0.03 |
ENSRNOT00000082894
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr17_-_44643362 | 0.03 |
ENSRNOT00000091041
|
Zfp184
|
zinc finger protein 184 |
| chr14_+_86414949 | 0.03 |
ENSRNOT00000083394
ENSRNOT00000057199 |
LOC103693780
|
2-oxoglutarate dehydrogenase, mitochondrial-like |
| chr4_-_100213858 | 0.03 |
ENSRNOT00000015562
|
Usp39
|
ubiquitin specific peptidase 39 |
| chr5_+_122100099 | 0.03 |
ENSRNOT00000007738
|
Pde4b
|
phosphodiesterase 4B |
| chr4_-_99822879 | 0.03 |
ENSRNOT00000012622
ENSRNOT00000078991 |
Ptcd3
|
Pentatricopeptide repeat domain 3 |
| chr19_+_37852659 | 0.03 |
ENSRNOT00000030967
|
Nrn1l
|
neuritin 1-like |
| chr3_+_122803772 | 0.03 |
ENSRNOT00000009564
|
Nop56
|
NOP56 ribonucleoprotein |
| chr12_-_52658275 | 0.03 |
ENSRNOT00000041981
|
Zfp605
|
zinc finger protein 605 |
| chr19_-_59894937 | 0.02 |
ENSRNOT00000027145
|
Rbm34
|
RNA binding motif protein 34 |
| chr3_-_8348746 | 0.02 |
ENSRNOT00000064638
|
Trub2
|
TruB pseudouridine synthase family member 2 |
| chr2_-_25095106 | 0.02 |
ENSRNOT00000051653
ENSRNOT00000085857 |
Aggf1
|
angiogenic factor with G patch and FHA domains 1 |
| chr2_-_59084059 | 0.02 |
ENSRNOT00000086323
ENSRNOT00000088701 |
Spef2
|
sperm flagellar 2 |
| chr16_+_70081035 | 0.02 |
ENSRNOT00000016098
|
LOC100912059
|
ubiquitin carboxyl-terminal hydrolase 12-like |
| chr2_-_192697800 | 0.02 |
ENSRNOT00000066614
|
NEWGENE_2324572
|
small proline rich protein 4 |
| chr10_+_65351245 | 0.02 |
ENSRNOT00000045107
|
Fam222b
|
family with sequence similarity 222, member B |
| chr1_-_67302751 | 0.02 |
ENSRNOT00000041518
|
Vom1r42
|
vomeronasal 1 receptor 42 |
| chr6_-_9459188 | 0.02 |
ENSRNOT00000019894
|
Srbd1
|
S1 RNA binding domain 1 |
| chrX_+_138046494 | 0.02 |
ENSRNOT00000010596
|
Stk26
|
serine/threonine kinase 26 |
| chr13_-_82298404 | 0.02 |
ENSRNOT00000087487
|
LOC498265
|
similar to hypothetical protein FLJ10706 |
| chr15_+_41927241 | 0.02 |
ENSRNOT00000012035
|
Trim13
|
tripartite motif-containing 13 |
| chr4_-_98305173 | 0.02 |
ENSRNOT00000010151
|
Il23r
|
interleukin 23 receptor |
| chr17_-_54714914 | 0.02 |
ENSRNOT00000024336
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr9_+_66952720 | 0.02 |
ENSRNOT00000023678
|
Nbeal1
|
neurobeachin-like 1 |
| chr9_+_52693831 | 0.02 |
ENSRNOT00000050275
|
Wdr75
|
WD repeat domain 75 |
| chr3_+_8349185 | 0.02 |
ENSRNOT00000038602
|
Coq4
|
coenzyme Q4 |
| chr19_-_23944820 | 0.02 |
ENSRNOT00000080337
|
Zfp330
|
zinc finger protein 330 |
| chr1_-_189460844 | 0.02 |
ENSRNOT00000020077
|
Thumpd1
|
THUMP domain containing 1 |
| chr1_-_87777668 | 0.02 |
ENSRNOT00000037288
|
AABR07002870.1
|
|
| chr13_+_98615287 | 0.02 |
ENSRNOT00000004032
|
Itpkb
|
inositol-trisphosphate 3-kinase B |
| chr7_+_131330913 | 0.02 |
ENSRNOT00000019672
|
Alg10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr1_-_191984858 | 0.02 |
ENSRNOT00000025309
|
Gga2
|
golgi associated, gamma adaptin ear containing, ARF binding protein 2 |
| chrX_+_99852198 | 0.02 |
ENSRNOT00000082451
|
AABR07040412.1
|
|
| chr3_+_47439076 | 0.02 |
ENSRNOT00000011794
|
Tank
|
TRAF family member-associated NFKB activator |
| chr3_-_12155098 | 0.02 |
ENSRNOT00000082696
|
Garnl3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr8_-_7426611 | 0.02 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr2_+_186980793 | 0.02 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr18_-_41389510 | 0.02 |
ENSRNOT00000005476
ENSRNOT00000005446 |
Sema6a
|
semaphorin 6A |
| chr4_-_17440506 | 0.01 |
ENSRNOT00000080931
|
Sema3e
|
semaphorin 3E |
| chr9_-_93377643 | 0.01 |
ENSRNOT00000024712
|
Ncl
|
nucleolin |
| chr2_+_116032455 | 0.01 |
ENSRNOT00000066495
|
Phc3
|
polyhomeotic homolog 3 |
| chr14_+_2100106 | 0.01 |
ENSRNOT00000000064
|
Gak
|
cyclin G associated kinase |
| chr9_-_17065600 | 0.01 |
ENSRNOT00000025638
|
Dlk2
|
delta like non-canonical Notch ligand 2 |
| chr15_-_33333417 | 0.01 |
ENSRNOT00000018982
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr20_+_41083317 | 0.01 |
ENSRNOT00000000660
|
Tspyl1
|
TSPY-like 1 |
| chrX_+_44907521 | 0.01 |
ENSRNOT00000004901
|
Tbl1x
|
transducin (beta)-like 1 X-linked |
| chr15_+_40937708 | 0.01 |
ENSRNOT00000018407
|
Spata13
|
spermatogenesis associated 13 |
| chr18_+_52215682 | 0.01 |
ENSRNOT00000037901
|
Megf10
|
multiple EGF-like domains 10 |
| chr4_+_31333970 | 0.01 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr14_+_43810119 | 0.01 |
ENSRNOT00000003327
|
Rbm47
|
RNA binding motif protein 47 |
| chrM_+_3904 | 0.01 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chrX_-_63182385 | 0.01 |
ENSRNOT00000076613
|
Zfx
|
zinc finger protein X-linked |
| chr16_+_39144972 | 0.01 |
ENSRNOT00000086728
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr11_-_4332255 | 0.01 |
ENSRNOT00000087133
|
Cadm2
|
cell adhesion molecule 2 |
| chr1_+_37507276 | 0.01 |
ENSRNOT00000047627
|
Adcy2
|
adenylate cyclase 2 |
| chr8_-_87282156 | 0.01 |
ENSRNOT00000087874
|
Filip1
|
filamin A interacting protein 1 |
| chr12_-_11175917 | 0.01 |
ENSRNOT00000079573
|
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr8_+_76754492 | 0.01 |
ENSRNOT00000085727
|
Myo1e
|
myosin IE |
| chr8_+_81949877 | 0.01 |
ENSRNOT00000030221
|
LOC100360828
|
cAMP-regulated phosphoprotein 19-like |
| chr14_+_36216002 | 0.01 |
ENSRNOT00000038506
|
Scfd2
|
sec1 family domain containing 2 |
| chr18_+_30474947 | 0.01 |
ENSRNOT00000027188
|
Pcdhb9
|
protocadherin beta 9 |
| chr2_+_196145384 | 0.01 |
ENSRNOT00000080966
|
Pi4kb
|
phosphatidylinositol 4-kinase beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmga2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:0090292 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0099526 | synaptic vesicle targeting(GO:0016080) presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.0 | GO:0046078 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) dUMP metabolic process(GO:0046078) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.0 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.0 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.0 | GO:0009384 | N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |