Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
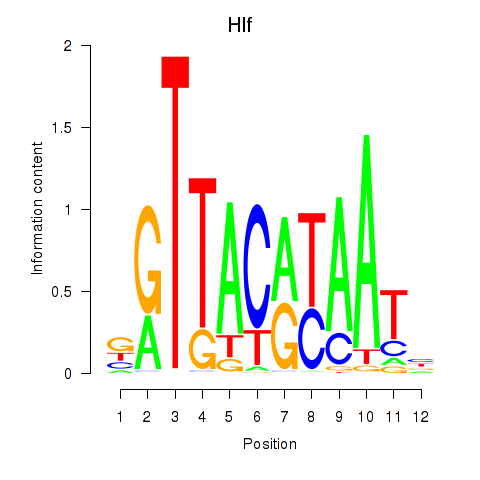
Results for Hlf
Z-value: 0.74
Transcription factors associated with Hlf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hlf
|
ENSRNOG00000002456 | HLF, PAR bZIP transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hlf | rn6_v1_chr10_-_77918191_77918191 | 0.17 | 7.8e-01 | Click! |
Activity profile of Hlf motif
Sorted Z-values of Hlf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_184188911 | 0.33 |
ENSRNOT00000055124
ENSRNOT00000014948 |
Calca
|
calcitonin-related polypeptide alpha |
| chr15_-_47442664 | 0.26 |
ENSRNOT00000072994
|
Prss55
|
protease, serine, 55 |
| chr12_-_35979193 | 0.22 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr5_-_168123395 | 0.22 |
ENSRNOT00000024932
|
Per3
|
period circadian clock 3 |
| chr15_+_87722221 | 0.21 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr11_-_27080701 | 0.21 |
ENSRNOT00000002180
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chrX_-_10218583 | 0.20 |
ENSRNOT00000013382
|
Nyx
|
nyctalopin |
| chr10_+_57239993 | 0.20 |
ENSRNOT00000067919
|
LOC687707
|
hypothetical protein LOC687707 |
| chr7_+_2435553 | 0.19 |
ENSRNOT00000065593
|
Prim1
|
primase (DNA) subunit 1 |
| chr17_+_11683862 | 0.19 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chrX_+_33884499 | 0.19 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr6_+_43743702 | 0.18 |
ENSRNOT00000082570
|
Grhl1
|
grainyhead-like transcription factor 1 |
| chr4_-_118472179 | 0.17 |
ENSRNOT00000023856
|
Mxd1
|
max dimerization protein 1 |
| chr2_+_196594303 | 0.17 |
ENSRNOT00000064442
ENSRNOT00000044738 |
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr6_+_144156175 | 0.16 |
ENSRNOT00000006582
|
Esyt2
|
extended synaptotagmin 2 |
| chr2_+_165805144 | 0.15 |
ENSRNOT00000073716
|
Rabif
|
RAB interacting factor |
| chr15_+_40665041 | 0.15 |
ENSRNOT00000018300
|
Amer2
|
APC membrane recruitment protein 2 |
| chr13_+_74456487 | 0.15 |
ENSRNOT00000065801
|
Angptl1
|
angiopoietin-like 1 |
| chr8_-_36764422 | 0.15 |
ENSRNOT00000017330
|
Hyls1
|
HYLS1, centriolar and ciliogenesis associated |
| chr12_+_4737817 | 0.15 |
ENSRNOT00000035992
|
AABR07035107.1
|
|
| chr18_-_36513317 | 0.15 |
ENSRNOT00000025453
|
Plac8l1
|
PLAC8-like 1 |
| chr1_-_61872975 | 0.14 |
ENSRNOT00000078809
|
AABR07001910.1
|
|
| chr9_+_73529612 | 0.14 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr5_-_7941822 | 0.14 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr10_-_66602987 | 0.14 |
ENSRNOT00000017949
|
Wsb1
|
WD repeat and SOCS box-containing 1 |
| chr10_-_16752205 | 0.13 |
ENSRNOT00000079462
|
Crebrf
|
CREB3 regulatory factor |
| chrX_-_29648359 | 0.13 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr4_-_161743847 | 0.13 |
ENSRNOT00000064174
ENSRNOT00000084173 ENSRNOT00000064215 |
Itfg2
|
integrin alpha FG-GAP repeat containing 2 |
| chr10_-_35187416 | 0.13 |
ENSRNOT00000084509
|
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr2_-_220838905 | 0.13 |
ENSRNOT00000078914
|
AABR07013065.1
|
|
| chr3_+_177310753 | 0.13 |
ENSRNOT00000031448
|
Myt1
|
myelin transcription factor 1 |
| chr2_-_247988462 | 0.13 |
ENSRNOT00000022387
|
Pdlim5
|
PDZ and LIM domain 5 |
| chr11_+_62584959 | 0.12 |
ENSRNOT00000071065
|
Gramd1c
|
GRAM domain containing 1C |
| chr8_+_49354115 | 0.12 |
ENSRNOT00000032837
|
Mpzl3
|
myelin protein zero-like 3 |
| chr12_-_5490935 | 0.12 |
ENSRNOT00000050885
|
Zfp958
|
zinc finger protein 958 |
| chrX_+_15113878 | 0.12 |
ENSRNOT00000007464
|
Wdr13
|
WD repeat domain 13 |
| chr16_+_54291251 | 0.12 |
ENSRNOT00000079006
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr1_+_28454966 | 0.12 |
ENSRNOT00000078841
ENSRNOT00000030327 |
Tpd52l1
|
tumor protein D52-like 1 |
| chr1_+_15834779 | 0.12 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr7_-_59587382 | 0.11 |
ENSRNOT00000091427
ENSRNOT00000066396 |
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr12_+_8725517 | 0.11 |
ENSRNOT00000001243
|
Slc46a3
|
solute carrier family 46, member 3 |
| chr3_+_95715193 | 0.11 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr7_+_63922879 | 0.11 |
ENSRNOT00000043581
|
RGD1565498
|
similar to Hypothetical protein LOC270802 |
| chr6_-_41870046 | 0.11 |
ENSRNOT00000005863
|
Lpin1
|
lipin 1 |
| chr12_-_29743705 | 0.11 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr12_+_38368693 | 0.11 |
ENSRNOT00000083261
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr18_+_40596166 | 0.11 |
ENSRNOT00000087984
|
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr10_-_47453442 | 0.10 |
ENSRNOT00000050061
|
Usp22
|
ubiquitin specific peptidase 22 |
| chr6_+_28515025 | 0.10 |
ENSRNOT00000088033
ENSRNOT00000005317 ENSRNOT00000081141 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr11_+_64882288 | 0.10 |
ENSRNOT00000077727
|
Pla1a
|
phospholipase A1 member A |
| chr6_+_99883959 | 0.10 |
ENSRNOT00000010588
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr4_-_22307453 | 0.10 |
ENSRNOT00000047126
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr18_+_30562178 | 0.10 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr8_+_52753011 | 0.10 |
ENSRNOT00000047264
|
Nxpe1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr10_+_106869401 | 0.10 |
ENSRNOT00000074369
|
Tmem235
|
transmembrane protein 235 |
| chr20_-_32501722 | 0.10 |
ENSRNOT00000000448
|
Zufsp
|
zinc finger with UFM1-specific peptidase domain |
| chr2_+_187990242 | 0.10 |
ENSRNOT00000092819
|
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr6_-_102472926 | 0.10 |
ENSRNOT00000079351
|
Zfyve26
|
zinc finger FYVE-type containing 26 |
| chr13_+_44475970 | 0.10 |
ENSRNOT00000024602
ENSRNOT00000091645 |
Ccnt2
|
cyclin T2 |
| chr11_-_1836897 | 0.09 |
ENSRNOT00000050533
|
Zfp654
|
zinc finger protein 654 |
| chr17_-_15566332 | 0.09 |
ENSRNOT00000093743
|
Ecm2
|
extracellular matrix protein 2 |
| chr1_-_247985497 | 0.09 |
ENSRNOT00000078998
|
Ranbp6
|
RAN binding protein 6 |
| chr7_+_66742987 | 0.09 |
ENSRNOT00000045130
|
Mon2
|
MON2 homolog, regulator of endosome-to-Golgi trafficking |
| chr1_-_70235091 | 0.09 |
ENSRNOT00000075745
|
Apeg3
|
antisense paternally expressed gene 3 |
| chr18_+_29966245 | 0.09 |
ENSRNOT00000074028
|
Pcdha4
|
protocadherin alpha 4 |
| chr8_+_116804451 | 0.09 |
ENSRNOT00000041402
|
Ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr6_+_64252970 | 0.09 |
ENSRNOT00000093700
|
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr1_+_217345545 | 0.09 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr18_+_14471213 | 0.09 |
ENSRNOT00000045224
|
Dtna
|
dystrobrevin, alpha |
| chr4_-_68597586 | 0.09 |
ENSRNOT00000015921
|
RGD1563986
|
similar to RIKEN cDNA E330009J07 gene |
| chr1_+_127604197 | 0.09 |
ENSRNOT00000018463
|
Lins1
|
lines homolog 1 |
| chr13_-_78957183 | 0.09 |
ENSRNOT00000076954
ENSRNOT00000003851 |
Klhl20
|
kelch-like family member 20 |
| chr8_+_69127708 | 0.09 |
ENSRNOT00000013490
|
Snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr1_-_56683731 | 0.08 |
ENSRNOT00000014552
|
Thbs2
|
thrombospondin 2 |
| chr17_+_85382116 | 0.08 |
ENSRNOT00000002513
|
Spag6
|
sperm associated antigen 6 |
| chr5_-_117440254 | 0.08 |
ENSRNOT00000066139
|
Kank4
|
KN motif and ankyrin repeat domains 4 |
| chr11_+_17538063 | 0.08 |
ENSRNOT00000031889
ENSRNOT00000090878 |
Chodl
|
chondrolectin |
| chr2_+_189629297 | 0.08 |
ENSRNOT00000049810
|
Dennd4b
|
DENN domain containing 4B |
| chr2_-_53300404 | 0.08 |
ENSRNOT00000088876
|
Ghr
|
growth hormone receptor |
| chr10_-_6870011 | 0.08 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
| chr4_-_85915099 | 0.08 |
ENSRNOT00000016182
|
Neurod6
|
neuronal differentiation 6 |
| chr14_+_17228856 | 0.08 |
ENSRNOT00000003082
|
Cxcl9
|
C-X-C motif chemokine ligand 9 |
| chrX_+_15064594 | 0.08 |
ENSRNOT00000007130
|
Tbc1d25
|
TBC1 domain family, member 25 |
| chr2_-_96668222 | 0.08 |
ENSRNOT00000016567
|
Pkia
|
cAMP-dependent protein kinase inhibitor alpha |
| chr15_-_2966576 | 0.08 |
ENSRNOT00000070893
ENSRNOT00000017383 |
Kat6b
|
lysine acetyltransferase 6B |
| chr7_-_117364697 | 0.08 |
ENSRNOT00000077314
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr3_-_146407252 | 0.08 |
ENSRNOT00000091310
|
Apmap
|
adipocyte plasma membrane associated protein |
| chr13_-_111972603 | 0.08 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr5_+_104394119 | 0.07 |
ENSRNOT00000093557
ENSRNOT00000093462 |
Adamtsl1
|
ADAMTS-like 1 |
| chrX_+_153064028 | 0.07 |
ENSRNOT00000079424
|
Pnma3
|
paraneoplastic Ma antigen 3 |
| chr4_+_70572942 | 0.07 |
ENSRNOT00000051964
|
AC142181.1
|
|
| chr7_-_120874308 | 0.07 |
ENSRNOT00000078320
|
Fam227a
|
family with sequence similarity 227, member A |
| chr16_+_54332660 | 0.07 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr11_-_17119582 | 0.07 |
ENSRNOT00000087665
|
RGD1563888
|
similar to DNA segment, Chr 16, ERATO Doi 472, expressed |
| chr5_+_58661049 | 0.07 |
ENSRNOT00000078274
|
Unc13b
|
unc-13 homolog B |
| chr17_+_7675531 | 0.07 |
ENSRNOT00000061187
|
Spock1
|
sparc/osteonectin, cwcv and kazal like domains proteoglycan 1 |
| chr6_-_107325345 | 0.07 |
ENSRNOT00000049481
ENSRNOT00000042594 ENSRNOT00000013026 |
Numb
|
NUMB, endocytic adaptor protein |
| chr8_-_132790778 | 0.07 |
ENSRNOT00000008255
ENSRNOT00000091431 |
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr13_-_98078555 | 0.07 |
ENSRNOT00000078034
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr4_+_157107469 | 0.07 |
ENSRNOT00000015678
|
C1rl
|
complement C1r subcomponent like |
| chr1_-_189666440 | 0.07 |
ENSRNOT00000019167
|
Dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr19_-_57333433 | 0.07 |
ENSRNOT00000024917
|
Agt
|
angiotensinogen |
| chr5_+_151181559 | 0.07 |
ENSRNOT00000085674
|
Fgr
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr20_-_2088389 | 0.07 |
ENSRNOT00000090910
|
Znrd1as1
|
ZNRD1 antisense RNA 1 |
| chr3_-_112371664 | 0.07 |
ENSRNOT00000079606
ENSRNOT00000012636 |
LOC100910478
|
leucine-rich repeat-containing protein 57-like |
| chr2_+_209661244 | 0.07 |
ENSRNOT00000091973
|
AABR07012826.1
|
|
| chr3_+_112371677 | 0.07 |
ENSRNOT00000013316
|
LOC100912076
|
HAUS augmin-like complex subunit 2-like |
| chr1_+_240355149 | 0.07 |
ENSRNOT00000018521
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr2_+_3662763 | 0.06 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr8_-_61519507 | 0.06 |
ENSRNOT00000038347
|
Odf3l1
|
outer dense fiber of sperm tails 3-like 1 |
| chr1_-_167308827 | 0.06 |
ENSRNOT00000027590
ENSRNOT00000027575 |
Nup98
|
nucleoporin 98 |
| chr8_+_44061894 | 0.06 |
ENSRNOT00000084416
|
Zfp202
|
zinc finger protein 202 |
| chr15_-_76789298 | 0.06 |
ENSRNOT00000077853
|
AABR07018857.1
|
|
| chr1_+_162768156 | 0.06 |
ENSRNOT00000049321
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr10_-_89700283 | 0.06 |
ENSRNOT00000028213
|
Etv4
|
ets variant 4 |
| chr18_+_79773608 | 0.06 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr1_-_242861767 | 0.06 |
ENSRNOT00000021185
|
Cbwd1
|
COBW domain containing 1 |
| chr5_+_169213115 | 0.06 |
ENSRNOT00000013535
|
Nol9
|
nucleolar protein 9 |
| chr1_-_156296161 | 0.06 |
ENSRNOT00000025653
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr18_+_45023932 | 0.06 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr2_+_247248407 | 0.06 |
ENSRNOT00000082287
|
Unc5c
|
unc-5 netrin receptor C |
| chr8_-_39190457 | 0.06 |
ENSRNOT00000090161
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr7_+_121311024 | 0.06 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr2_-_152824547 | 0.06 |
ENSRNOT00000019824
|
Dhx36
|
DEAH-box helicase 36 |
| chr1_+_27924825 | 0.06 |
ENSRNOT00000087830
|
AABR07000902.1
|
|
| chr6_+_101924000 | 0.06 |
ENSRNOT00000092157
|
Mpp5
|
membrane palmitoylated protein 5 |
| chr14_-_14192048 | 0.06 |
ENSRNOT00000060868
|
Bmp2k
|
BMP2 inducible kinase |
| chr9_+_67234303 | 0.06 |
ENSRNOT00000050179
|
Abi2
|
abl-interactor 2 |
| chr19_+_37990374 | 0.06 |
ENSRNOT00000026827
|
Dus2
|
dihydrouridine synthase 2 |
| chr5_+_120340646 | 0.06 |
ENSRNOT00000086259
ENSRNOT00000086539 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr14_-_34561696 | 0.06 |
ENSRNOT00000059763
|
Srd5a3
|
steroid 5 alpha-reductase 3 |
| chr4_-_18396035 | 0.06 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr5_-_164679654 | 0.06 |
ENSRNOT00000035695
|
LOC100911456
|
migration and invasion-inhibitory protein-like |
| chr13_-_84759098 | 0.06 |
ENSRNOT00000058683
|
LOC685351
|
similar to flavin containing monooxygenase 5 |
| chr16_+_65256751 | 0.06 |
ENSRNOT00000092012
|
AABR07026137.2
|
|
| chr9_+_118586179 | 0.06 |
ENSRNOT00000022351
|
Dlgap1
|
DLG associated protein 1 |
| chr11_+_42259761 | 0.05 |
ENSRNOT00000047310
|
Epha6
|
Eph receptor A6 |
| chr5_-_50068706 | 0.05 |
ENSRNOT00000084643
|
Orc3
|
origin recognition complex, subunit 3 |
| chr20_+_10844266 | 0.05 |
ENSRNOT00000001583
|
Rrp1b
|
ribosomal RNA processing 1B |
| chr2_+_252771017 | 0.05 |
ENSRNOT00000075452
ENSRNOT00000055318 |
Ttll7
|
tubulin tyrosine ligase like 7 |
| chr13_-_98078946 | 0.05 |
ENSRNOT00000035875
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chrX_+_158978755 | 0.05 |
ENSRNOT00000066809
|
Slc9a6
|
solute carrier family 9 member A6 |
| chr10_-_89699836 | 0.05 |
ENSRNOT00000084311
|
Etv4
|
ets variant 4 |
| chr10_-_90030639 | 0.05 |
ENSRNOT00000090456
|
Mpp2
|
membrane palmitoylated protein 2 |
| chr2_-_164684985 | 0.05 |
ENSRNOT00000057504
|
Rarres1
|
retinoic acid receptor responder 1 |
| chr8_+_99636749 | 0.05 |
ENSRNOT00000086524
|
Plscr1
|
phospholipid scramblase 1 |
| chr1_-_247088012 | 0.05 |
ENSRNOT00000020464
|
Spata6l
|
spermatogenesis associated 6-like |
| chr3_+_176989672 | 0.05 |
ENSRNOT00000020373
|
Tpd52l2
|
tumor protein D52-like 2 |
| chr2_-_43067956 | 0.05 |
ENSRNOT00000090616
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chrX_+_137197396 | 0.05 |
ENSRNOT00000082227
|
RGD1566225
|
similar to RIKEN cDNA 1700001F22 |
| chr2_-_30340103 | 0.05 |
ENSRNOT00000024023
ENSRNOT00000067722 |
Bdp1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr18_-_77322690 | 0.05 |
ENSRNOT00000058382
|
Nfatc1
|
nuclear factor of activated T-cells 1 |
| chr20_+_22728208 | 0.05 |
ENSRNOT00000000794
|
LOC100363472
|
nuclear receptor-binding factor 2-like |
| chr19_+_15195565 | 0.05 |
ENSRNOT00000090865
ENSRNOT00000078874 |
Ces1d
|
carboxylesterase 1D |
| chr18_-_74198369 | 0.05 |
ENSRNOT00000073597
|
LOC108348136
|
HAUS augmin-like complex subunit 1 |
| chr5_-_128446725 | 0.05 |
ENSRNOT00000011272
|
Btf3l4
|
basic transcription factor 3-like 4 |
| chr7_-_59547174 | 0.05 |
ENSRNOT00000085836
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr11_+_61761613 | 0.05 |
ENSRNOT00000089340
ENSRNOT00000091598 |
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr2_-_1561464 | 0.05 |
ENSRNOT00000062055
ENSRNOT00000062054 ENSRNOT00000062052 |
Cast
|
calpastatin |
| chr1_-_209641123 | 0.05 |
ENSRNOT00000021702
|
Ebf3
|
early B-cell factor 3 |
| chr4_+_152430509 | 0.05 |
ENSRNOT00000076062
|
Rad52
|
RAD52 homolog, DNA repair protein |
| chr1_+_227592635 | 0.05 |
ENSRNOT00000072135
|
Ms4a4a
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr10_-_89084885 | 0.05 |
ENSRNOT00000027452
|
Plekhh3
|
pleckstrin homology, MyTH4 and FERM domain containing H3 |
| chr3_-_164964702 | 0.05 |
ENSRNOT00000014595
|
Adnp
|
activity-dependent neuroprotector homeobox |
| chr2_+_188052236 | 0.05 |
ENSRNOT00000027385
|
RGD1565775
|
similar to RIKEN cDNA 2810403A07 |
| chr10_-_87954055 | 0.04 |
ENSRNOT00000018048
|
Krt34
|
keratin 34 |
| chr3_+_124515978 | 0.04 |
ENSRNOT00000028881
|
Prnp
|
prion protein |
| chr10_-_78156477 | 0.04 |
ENSRNOT00000083009
|
Stxbp4
|
syntaxin binding protein 4 |
| chr9_+_114022137 | 0.04 |
ENSRNOT00000007657
|
Map3k7
|
mitogen activated protein kinase kinase kinase 7 |
| chr12_+_38965063 | 0.04 |
ENSRNOT00000074936
|
Morn3
|
MORN repeat containing 3 |
| chr20_+_5933303 | 0.04 |
ENSRNOT00000000617
|
Mapk14
|
mitogen activated protein kinase 14 |
| chr1_+_101603222 | 0.04 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr7_-_142180794 | 0.04 |
ENSRNOT00000037447
|
Tfcp2
|
transcription factor CP2 |
| chr7_-_139063752 | 0.04 |
ENSRNOT00000072309
|
LOC102551901
|
protein lifeguard 2-like |
| chr2_+_40554146 | 0.04 |
ENSRNOT00000015138
|
Pde4d
|
phosphodiesterase 4D |
| chr11_-_79703736 | 0.04 |
ENSRNOT00000044279
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr5_+_76386836 | 0.04 |
ENSRNOT00000088872
ENSRNOT00000021110 |
Ugcg
|
UDP-glucose ceramide glucosyltransferase |
| chr1_+_30863217 | 0.04 |
ENSRNOT00000015421
|
Rnf146
|
ring finger protein 146 |
| chr3_-_112385528 | 0.04 |
ENSRNOT00000049756
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr2_+_251200686 | 0.04 |
ENSRNOT00000019210
|
Col24a1
|
collagen type XXIV alpha 1 chain |
| chr7_-_2941122 | 0.04 |
ENSRNOT00000082107
|
Esyt1
|
extended synaptotagmin 1 |
| chr6_-_125723732 | 0.04 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr10_+_45559578 | 0.04 |
ENSRNOT00000004011
|
RGD1304587
|
similar to RIKEN cDNA 2310033P09 |
| chr4_-_168517177 | 0.04 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
| chr17_-_76188979 | 0.04 |
ENSRNOT00000092442
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr5_-_100970043 | 0.04 |
ENSRNOT00000013950
|
Zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr2_-_192671059 | 0.04 |
ENSRNOT00000012174
|
Sprr1a
|
small proline-rich protein 1A |
| chr18_-_1828606 | 0.04 |
ENSRNOT00000033312
|
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr7_-_142180997 | 0.04 |
ENSRNOT00000087632
|
Tfcp2
|
transcription factor CP2 |
| chr7_+_130308532 | 0.04 |
ENSRNOT00000011941
|
Miox
|
myo-inositol oxygenase |
| chr18_-_28871849 | 0.04 |
ENSRNOT00000025902
|
Nrg2
|
neuregulin 2 |
| chr5_+_148577332 | 0.04 |
ENSRNOT00000016325
|
Snrnp40
|
small nuclear ribonucleoprotein U5 subunit 40 |
| chr2_-_57935334 | 0.04 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr13_+_105684420 | 0.03 |
ENSRNOT00000040543
|
Gpatch2
|
G patch domain containing 2 |
| chr2_-_22096949 | 0.03 |
ENSRNOT00000089325
|
Zfyve16
|
zinc finger FYVE-type containing 16 |
| chr17_+_507377 | 0.03 |
ENSRNOT00000023638
|
Npepo
|
aminopeptidase O |
| chr1_-_62558033 | 0.03 |
ENSRNOT00000015520
|
Zfp40
|
zinc finger protein 40 |
| chr3_+_148635775 | 0.03 |
ENSRNOT00000067261
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr8_+_114866768 | 0.03 |
ENSRNOT00000076731
|
Wdr82
|
WD repeat domain 82 |
| chr9_+_73378057 | 0.03 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hlf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 0.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:1905231 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 0.0 | 0.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:1903598 | ovarian follicle rupture(GO:0001543) angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of renal output by angiotensin(GO:0002019) angiotensin-mediated drinking behavior(GO:0003051) regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.2 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0021888 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.0 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.2 | GO:0098880 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.0 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.0 | GO:0044317 | rod spherule(GO:0044317) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0052723 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |