Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
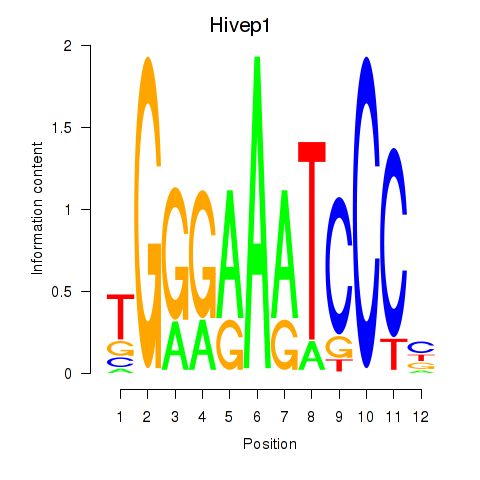
Results for Hivep1
Z-value: 0.16
Transcription factors associated with Hivep1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hivep1
|
ENSRNOG00000014460 | human immunodeficiency virus type I enhancer binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hivep1 | rn6_v1_chr17_-_22311392_22311392 | -0.30 | 6.2e-01 | Click! |
Activity profile of Hivep1 motif
Sorted Z-values of Hivep1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_76270457 | 0.10 |
ENSRNOT00000009894
|
Nfkbia
|
NFKB inhibitor alpha |
| chr12_-_17186679 | 0.10 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chrX_+_111122552 | 0.05 |
ENSRNOT00000083566
ENSRNOT00000085078 ENSRNOT00000090928 |
Cldn2
|
claudin 2 |
| chr20_-_4921348 | 0.05 |
ENSRNOT00000082497
ENSRNOT00000041151 |
RT1-CE4
|
RT1 class I, locus CE4 |
| chr12_+_24978483 | 0.04 |
ENSRNOT00000040069
|
Eln
|
elastin |
| chr18_+_32273770 | 0.03 |
ENSRNOT00000087408
|
Fgf1
|
fibroblast growth factor 1 |
| chr18_+_57011575 | 0.03 |
ENSRNOT00000026679
|
Il17b
|
interleukin 17B |
| chr4_-_93406182 | 0.03 |
ENSRNOT00000007437
|
AABR07060778.1
|
|
| chr14_-_18733391 | 0.03 |
ENSRNOT00000003745
|
Cxcl2
|
C-X-C motif chemokine ligand 2 |
| chr20_-_4863011 | 0.03 |
ENSRNOT00000079503
|
Ltb
|
lymphotoxin beta |
| chr13_+_111870121 | 0.03 |
ENSRNOT00000007333
|
Irf6
|
interferon regulatory factor 6 |
| chr5_+_48011864 | 0.03 |
ENSRNOT00000067610
|
Mdn1
|
midasin AAA ATPase 1 |
| chr3_-_176958880 | 0.03 |
ENSRNOT00000078661
|
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr9_+_9842585 | 0.03 |
ENSRNOT00000070806
|
Cd70
|
Cd70 molecule |
| chr8_+_59900651 | 0.02 |
ENSRNOT00000020410
|
Tmem266
|
transmembrane protein 266 |
| chr1_-_219745654 | 0.02 |
ENSRNOT00000054848
|
LOC689065
|
hypothetical protein LOC689065 |
| chr20_+_5184515 | 0.02 |
ENSRNOT00000089411
|
LOC103694381
|
lymphotoxin-beta |
| chr1_-_167347662 | 0.02 |
ENSRNOT00000027641
ENSRNOT00000076592 |
Rhog
|
ras homolog family member G |
| chr20_-_4863198 | 0.02 |
ENSRNOT00000001108
|
Ltb
|
lymphotoxin beta |
| chr13_+_56096834 | 0.02 |
ENSRNOT00000035129
|
Dennd1b
|
DENN domain containing 1B |
| chr3_-_7278758 | 0.02 |
ENSRNOT00000017004
|
Spaca9
|
sperm acrosome associated 9 |
| chr4_-_115239723 | 0.02 |
ENSRNOT00000042699
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr9_-_11114843 | 0.02 |
ENSRNOT00000072031
|
Ebi3
|
Epstein-Barr virus induced 3 |
| chr1_-_252550394 | 0.02 |
ENSRNOT00000083468
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr4_+_153217782 | 0.02 |
ENSRNOT00000015499
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr2_-_9023104 | 0.02 |
ENSRNOT00000039652
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr10_+_88764732 | 0.02 |
ENSRNOT00000026662
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr4_-_163049084 | 0.02 |
ENSRNOT00000091644
|
Cd69
|
Cd69 molecule |
| chr2_+_60169517 | 0.01 |
ENSRNOT00000080974
|
Prlr
|
prolactin receptor |
| chr1_+_86948918 | 0.01 |
ENSRNOT00000084839
|
Sirt2
|
sirtuin 2 |
| chr5_+_98469047 | 0.01 |
ENSRNOT00000041374
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr4_-_7228675 | 0.01 |
ENSRNOT00000064386
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr1_-_86948845 | 0.01 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chr9_-_64096265 | 0.01 |
ENSRNOT00000013502
|
Tyw5
|
tRNA-yW synthesizing protein 5 |
| chr7_+_144078496 | 0.01 |
ENSRNOT00000055302
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr4_-_145454834 | 0.01 |
ENSRNOT00000013154
ENSRNOT00000056493 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr1_-_141471010 | 0.01 |
ENSRNOT00000082164
|
Plin1
|
perilipin 1 |
| chr3_-_93216495 | 0.01 |
ENSRNOT00000010580
|
Ehf
|
ets homologous factor |
| chr5_-_139749050 | 0.01 |
ENSRNOT00000056651
|
Nfyc
|
nuclear transcription factor Y subunit gamma |
| chr20_+_2057689 | 0.01 |
ENSRNOT00000086007
|
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr16_-_50501921 | 0.01 |
ENSRNOT00000081023
|
Fat1
|
FAT atypical cadherin 1 |
| chr3_+_138715570 | 0.01 |
ENSRNOT00000064723
|
Sec23b
|
Sec23 homolog B, coat complex II component |
| chr1_+_166433109 | 0.01 |
ENSRNOT00000026428
|
Stard10
|
StAR-related lipid transfer domain containing 10 |
| chr9_-_4447715 | 0.01 |
ENSRNOT00000061882
|
Kat2b
|
lysine acetyltransferase 2B |
| chr10_+_86860685 | 0.01 |
ENSRNOT00000084644
|
Rara
|
retinoic acid receptor, alpha |
| chr12_+_40553741 | 0.01 |
ENSRNOT00000057396
|
LOC100360238
|
rCG21419-like |
| chr13_+_112031594 | 0.01 |
ENSRNOT00000008440
|
Lamb3
|
laminin subunit beta 3 |
| chr1_+_219764001 | 0.01 |
ENSRNOT00000082388
|
Pc
|
pyruvate carboxylase |
| chr1_+_84470829 | 0.01 |
ENSRNOT00000025472
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr17_+_5311274 | 0.01 |
ENSRNOT00000067020
|
LOC102547665
|
spermatogenesis-associated protein 31D1-like |
| chr20_-_4896970 | 0.01 |
ENSRNOT00000061114
ENSRNOT00000060980 |
RT1-CE7
RT1-CE5
|
RT1 class I, locus CE7 RT1 class I, locus CE5 |
| chr10_+_63803309 | 0.01 |
ENSRNOT00000036666
|
Myo1c
|
myosin 1C |
| chr20_+_2057878 | 0.01 |
ENSRNOT00000051480
|
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr4_-_157266018 | 0.01 |
ENSRNOT00000019570
|
LOC100911713
|
protein C10-like |
| chr4_+_153330069 | 0.01 |
ENSRNOT00000015560
|
Slc25a18
|
solute carrier family 25 member 18 |
| chr6_-_33507542 | 0.01 |
ENSRNOT00000007928
|
Gdf7
|
growth differentiation factor 7 |
| chr15_+_28377996 | 0.01 |
ENSRNOT00000078475
|
Arhgef40
|
Rho guanine nucleotide exchange factor 40 |
| chr9_+_112360419 | 0.01 |
ENSRNOT00000086682
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr20_+_3555135 | 0.01 |
ENSRNOT00000085380
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_-_87777359 | 0.01 |
ENSRNOT00000074090
|
AABR07002870.1
|
|
| chr17_+_11101306 | 0.01 |
ENSRNOT00000030893
|
Drd1
|
dopamine receptor D1 |
| chr20_-_4698718 | 0.01 |
ENSRNOT00000047527
|
RT1-CE7
|
RT1 class I, locus CE7 |
| chr5_+_168019058 | 0.01 |
ENSRNOT00000077202
|
Tnfrsf9
|
TNF receptor superfamily member 9 |
| chr1_-_183630695 | 0.01 |
ENSRNOT00000036330
|
AABR07005506.1
|
|
| chr18_-_62476700 | 0.01 |
ENSRNOT00000048429
|
Cycs
|
cytochrome c, somatic |
| chr1_+_164502389 | 0.01 |
ENSRNOT00000043554
|
Arrb1
|
arrestin, beta 1 |
| chr1_+_252589785 | 0.01 |
ENSRNOT00000025928
|
Fas
|
Fas cell surface death receptor |
| chr10_-_36502313 | 0.01 |
ENSRNOT00000041273
|
LOC679794
|
similar to Cytochrome c, somatic |
| chr20_-_3796258 | 0.01 |
ENSRNOT00000048076
ENSRNOT00000044520 |
RT1-CE16
|
RT1 class I, locus CE16 |
| chr3_+_95233874 | 0.01 |
ENSRNOT00000079743
|
LOC691083
|
hypothetical protein LOC691083 |
| chr7_+_116745061 | 0.01 |
ENSRNOT00000076174
|
Rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr1_-_220136470 | 0.01 |
ENSRNOT00000026812
|
Actn3
|
actinin alpha 3 |
| chr4_+_78080310 | 0.01 |
ENSRNOT00000035906
|
Sspo
|
SCO-spondin |
| chr1_+_199217016 | 0.01 |
ENSRNOT00000025732
|
Orai3
|
ORAI calcium release-activated calcium modulator 3 |
| chr8_-_127782070 | 0.00 |
ENSRNOT00000045493
|
Plcd1
|
phospholipase C, delta 1 |
| chrX_+_122724129 | 0.00 |
ENSRNOT00000017746
|
LOC100360218
|
interleukin 13 receptor, alpha 1-like |
| chr5_+_25146949 | 0.00 |
ENSRNOT00000061370
|
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr18_+_80939875 | 0.00 |
ENSRNOT00000021729
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr4_+_71790701 | 0.00 |
ENSRNOT00000085902
|
Tas2r143
|
taste receptor, type 2, member 143 |
| chr17_+_88963981 | 0.00 |
ENSRNOT00000025053
|
Myo3a
|
myosin IIIA |
| chr8_-_39093277 | 0.00 |
ENSRNOT00000043344
|
LOC100911068
|
roundabout homolog 4-like |
| chr5_+_133864798 | 0.00 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr20_-_157665 | 0.00 |
ENSRNOT00000048858
ENSRNOT00000079494 |
RT1-CE10
|
RT1 class I, locus CE10 |
| chr10_-_72196437 | 0.00 |
ENSRNOT00000029769
|
Pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr10_-_88645364 | 0.00 |
ENSRNOT00000030344
|
Rab5c
|
RAB5C, member RAS oncogene family |
| chr14_+_43810119 | 0.00 |
ENSRNOT00000003327
|
Rbm47
|
RNA binding motif protein 47 |
| chr9_-_93607087 | 0.00 |
ENSRNOT00000025262
|
Pde6d
|
phosphodiesterase 6D |
| chr12_-_22680630 | 0.00 |
ENSRNOT00000041808
|
Vgf
|
VGF nerve growth factor inducible |
| chr7_-_125795980 | 0.00 |
ENSRNOT00000017420
|
Arhgap8
|
Rho GTPase activating protein 8 |
| chr15_+_34552410 | 0.00 |
ENSRNOT00000027802
|
Khnyn
|
KH and NYN domain containing |
| chr5_-_107858104 | 0.00 |
ENSRNOT00000092196
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B |
| chrX_+_43831892 | 0.00 |
ENSRNOT00000005120
|
Myl6b
|
myosin light chain 6B |
| chr3_+_137618898 | 0.00 |
ENSRNOT00000007249
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr2_+_188253378 | 0.00 |
ENSRNOT00000085690
|
Ash1l
|
ASH1 like histone lysine methyltransferase |
| chr8_-_77398156 | 0.00 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr18_-_32207749 | 0.00 |
ENSRNOT00000090882
ENSRNOT00000018843 |
Arhgap26
|
Rho GTPase activating protein 26 |
| chr17_+_77167740 | 0.00 |
ENSRNOT00000042881
|
Optn
|
optineurin |
| chr10_-_43931094 | 0.00 |
ENSRNOT00000039778
|
Olr1415
|
olfactory receptor 1415 |
| chr2_-_142885604 | 0.00 |
ENSRNOT00000031487
|
Frem2
|
Fras1 related extracellular matrix protein 2 |
| chr2_-_187372652 | 0.00 |
ENSRNOT00000025496
ENSRNOT00000083116 |
Bcan
|
brevican |
| chr6_-_128727374 | 0.00 |
ENSRNOT00000082152
|
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr10_+_57278307 | 0.00 |
ENSRNOT00000005612
|
Eno3
|
enolase 3 |
| chr2_-_102871257 | 0.00 |
ENSRNOT00000013116
|
Cyp7b1
|
cytochrome P450, family 7, subfamily b, polypeptide 1 |
| chr7_+_14529482 | 0.00 |
ENSRNOT00000090844
ENSRNOT00000060284 ENSRNOT00000093239 |
Cyp4f5
Cyp4f17
|
cytochrome P450, family 4, subfamily f, polypeptide 5 cytochrome P450, family 4, subfamily f, polypeptide 17 |
| chr8_-_119265157 | 0.00 |
ENSRNOT00000056100
|
Rtp3
|
receptor (chemosensory) transporter protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hivep1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |